Head and neck squamous cell carcinomas (HNSCCs) encompass a diverse group of tumours arising from the nasopharynx, oral cavity, oropharynx, hypopharynx and larynx. Tobacco and alcohol abuse are risk factors for the development of HNSCC, particularly oral cavity, hypopharyngeal and laryngeal cancers, whereas Epstein–Barr virus (EBV) and human papillomavirus (HPV) can mediate tumourigenesis in nasopharyngeal carcinomas and oropharyngeal squamous cell carcinomas (OPSCCs), respectively.1,2 Head and neck cancers driven by HPV infection comprise a subgroup of tumours with distinct epidemiological and clinical characteristics. Compared with patients with HPV-negative HNSCC, patients with HPV-positive HNSCCs are typically younger than 60 years, male and white, with lower rates of tobacco use and exposure to higher numbers of oral sex partners.3 However, despite the decline in tobacco use in the past 20 years and the associated reduction in HNSCC incidence, the incidence of HPV-related OPSCC continues to rise worldwide, especially in developed countries.4 In the USA, the percentage of HPV prevalence in OPSCC increased from 16% in the 1980s to 72% during the early 2000s.5 Of the various HPV subtypes, HPV16 accounts for approximately 90% of cases of HPV-positive HNSCCs.6
Tumour HPV status is an independent prognostic factor in patients with OPSCC. HPV-positive OPSCCs are associated with higher response rates after induction chemotherapy and chemoradiation and with improved overall survival compared with HPV-negative OPSCCs.6–9 Given the impact of HPV status on prognosis in OPSCC, the American Joint Committee on Cancer staging system was updated to distinguish the staging for HPV-positive OPSCCs from that of HPV-negative HNSCCs.10 Although prognostic factors such as tumour HPV status have been established, there is a need in HNSCC to identify predictive biomarkers. Programmed death-ligand 1 is currently the only predictive biomarker in head and neck cancer, identifying survival benefits with pembrolizumab over chemotherapy in patients with metastatic or recurrent disease and high programmed death-ligand 1 combined positive score.11
Given the advancements in molecular technology, cell-free circulating DNA (cfDNA) has emerged with a range of potential applications in cancer medicine. Circulating tumour DNA (ctDNA) are fragments of cfDNA in the peripheral blood that contain specific genomic sequences derived from tumour tissue. ctDNA testing has the advantage of being minimally invasive, and detectable viral cfDNA in patients with virally driven tumours, such as nasopharyngeal carcinoma and OPSCC, is likely to be cancer specific.12 In nasopharyngeal carcinoma, detectable plasma EBV DNA after radiation correlates with an increased risk for locoregional failure, distant metastasis and death.13 Additionally, detectable plasma EBV DNA during radiation in advanced nasopharyngeal carcinoma is an adverse prognosticator of treatment outcomes.14 Studies in HPV-positive head and neck cancers suggest the promise of HPV ctDNA in the early detection and monitoring of disease and in predicting response to therapy.15 In this article, we review the current evidence and utility of HPV ctDNA in HNSCC.
Analysis of human papillomavirus circulating tumour DNA
HPV induces tumourigenesis by integrating its viral oncogenes into the somatic genome, thereby allowing the molecular detection of HPV ctDNA in primary tumours and bodily fluids.16 The HPV E6 and E7 oncoproteins are largely responsible for carcinogenesis through the inactivation of p53- and retinoblastoma-mediated pathways.17 Assays used to measure HPV ctDNA most commonly assess for E6 and/or E7 genes and include quantitative polymerase chain reaction (PCR) (qPCR), droplet digital PCR (ddPCR) and next-generation sequencing (NGS).18 Analyses by qPCR rely on specific primers to amplify target genes and report relative DNA quantification. ddPCR provides absolute quantification of DNA by fractionating samples into thousands of droplets containing DNA and subsequently amplifies each separated sample, resulting in high sensitivity and specificity. NGS can test for multiple genes at the same time, allowing for the detection of various HPV genotypes.15 Studies to date have primarily evaluated the utility of qPCR or ddPCR in analysing HPV ctDNA in the plasma, serum and saliva of patients with head and neck cancers (Table 1).19–35
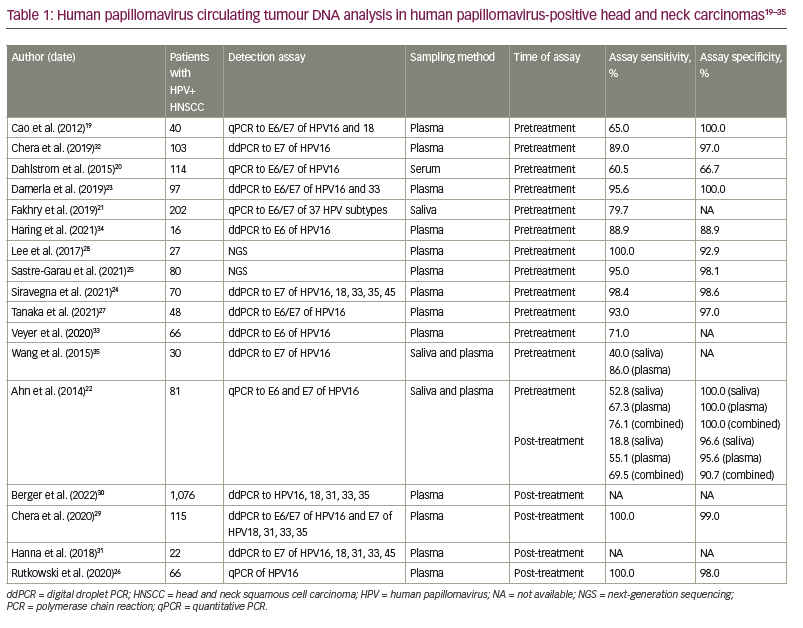
Over the years, numerous assays have been developed to measure HPV ctDNA in patients with HPV-positive head and neck carcinoma with varying sensitivity and specificity. One of the early studies using qPCR, Cao et al., found detectable HPV ctDNA in 65% of the pretreatment plasma samples of 40 patients with HPV-positive oropharyngeal carcinoma.19 Overall, the sensitivity of qPCR in detecting baseline HPV ctDNA has been modest, only reaching approximately 80%.19–22 Advancements in ddPCR and NGS technology, meanwhile, enabled these platforms to measure HPV ctDNA with greater accuracy. ddPCR can detect pretreatment plasma HPV ctDNA in patients with HPV-positive HNSCC with sensitivities and specificities of over 95%.23,24 A study by Sastre-Garau et al. using NGS demonstrated a sensitivity of 95% and specificity of 98% for detecting HPV ctDNA in pretreatment plasma.25 Notably, NGS exhibits an ultrasensitive ability to quantify HPV ctDNA. Sloane et al. described the ability of NGS to detect HPV ctDNA in pretreatment plasma samples at levels as low as 1 copy/mL, while Leung et al. detected HPV ctDNA in ddPCR-negative post-treatment samples of patients with cervical and oropharynx cancers at levels as low as 0.03 copies/mL plasma by NGS.36,37
Pretreatment human papillomavirus circulating tumour DNA analysis: Screening and diagnosis
Unlike breast, cervical, colon and lung cancers, there are currently no screening guidelines for the early detection of HNSCCs. The analysis of plasma EBV DNA has been found to be useful in screening for early nasopharyngeal carcinoma in patients at high risk for this disease; testing for circulating plasma EBV DNA by qPCR in asymptomatic Chinese patients was associated with a higher proportion of nasopharyngeal carcinoma diagnosed at earlier stages and an improved 3-year progression-free survival.38
HPV cfDNA analysis is a conceivable modality in the early diagnosis of HPV-positive HNSCCs.23,39,40 HPV16-E6 antibody seroconversion pre-dates an OPSCC diagnosis, with a median time of 11.5 years.41 OPSCC HPV16-E6 seropositivity confers an estimated 10-year risk for OPSCC of 17–27% for men and 4–6% for women aged 50–60 years; conversely, the estimated 10-year risk for OPSCC for seronegative individuals is 0.01–0.25%.40 Given the elevated risk for OPSCC associated with HPV16-E6 seropositivity, a screening strategy that incorporates periodic surveillance in high-risk HPV-seropositive individuals could be considered. One approach could implement minimally invasive serial biomarker testing with HPV ctDNA.40 A small study of 10 patients with HPV-positive HNSCC found detectable HPV ctDNA in three patients (30%) at 19, 34 and 43 months preceding an OPSCC diagnosis, demonstrating the presence of detectable HPV ctDNA in a subgroup of patients years before a clinical diagnosis of cancer.39
Current approaches to diagnosing HPV-positive HNSCCs necessitate tissue biopsy and assessment of HPV tumour status via p16 expression by immunohistochemistry. However, the specificity of p16 immunohistochemistry is only fairly optimal, with a false-positive rate of 3.8–7.3%.42,43 Siravegna et al. demonstrated the potential of HPV ctDNA as a non-invasive and accurate technique in diagnosing HPV-positive HNSCC.24 Using ddPCR assays for HPV genotypes 16, 18, 33, 35 and 45, the sensitivity and specificity of HPV ctDNA in detecting HPV-positive HNSCC were found to be 98.4% and 98.6%, respectively. Median time to diagnosis decreased by 63% or 26 days using a non-invasive diagnostic approach that included HPV ctDNA analysis compared with standard diagnostic approaches.24 HPV ctDNA analysis as a method of diagnosis for HPV-positive HNSCC is promising, with data by Siravegna et al. highlighting its feasibility, accuracy and time to diagnosis.24 HPV ctDNA as a diagnostic approach requires further validation in large prospective studies.
Post-treatment human papillomavirus circulating tumour DNA analysis: Detecting residual disease and surveillance
Studies suggest an adjunctive role for HPV ctDNA testing with imaging following definitive therapy for HPV-positive HNSCC.26–28 Given the suboptimal positive predictive value (PPV) of post-treatment positron emission tomography–computed tomography (PET-CT) scans, combining HPV ctDNA analysis and imaging may enhance the detection of residual disease.44 A prospective study of 35 patients by Tanaka et al. showed similar negative predictive values (NPV) for surveillance HPV ctDNA and PET-CT scans (89.7% versus 84.0%), whereas the PPV for HPV ctDNA was significantly higher than PET-CT scans (100% versus 50.0%).27 Lee et al. evaluated plasma HPV ctDNA in the post-treatment setting following chemoradiotherapy using NGS in patients with locally advanced HPV-positive HNSCC.28 Specimens from three patients who underwent post-treatment biopsy for increased 18F-fluorodeoxyglucose (18F-FDG) uptake in the primary site and three patients who underwent neck dissection for increased 18F-FDG uptake in cervical lymph nodes demonstrated no residual tumour, and corresponding plasma HPV ctDNA were undetectable in all patients. Hence, these studies shed light on the potential of HPV ctDNA as a biomarker of residual disease, avoiding unnecessary biopsies and surgical procedures despite positive PET-scan findings after definitive chemoradiation.28
Currently, the National Comprehensive Cancer Network guidelines recommend regular clinical head and neck examinations and fibreoptic evaluations for the surveillance of patients with non-metastatic HNSCC following definitive therapy.45 HPV ctDNA analysis is sensitive in detecting recurrence with promise for clinical application in surveillance.29,30 One retrospective study by Berger et al. evaluated the utility of HPV ctDNA as a biomarker of recurrent disease in 1,076 patients with HPV-positive OPSCC.30 Eighty patients (7.4%) had at least one positive HPV ctDNA. By using ddPCR, HPV ctDNA analysis demonstrated a PPV of 95% for disease recurrence and an NPV of 95%. Of the patients with positive surveillance HPV ctDNA tests who were not known to have disease recurrence by clinical or imaging examinations, the majority of patients (93%) were later confirmed to have recurrent disease, illustrating the capacity of HPV ctDNA to detect occult malignancy.30 A prospective study by Chera et al. corroborated the encouraging evidence for HPV ctDNA surveillance, demonstrating a PPV of 94% and NPV of 100% with consecutive plasma sampling and earlier detection of recurrence by a median time of 3.9 months before biopsy-confirmed recurrence.29
Prognostication and prediction of clinical outcomes
Since the emergence of ctDNA technology, efforts have been made to understand the clinical significance of HPV ctDNA in HNSCC. HPV ctDNA can inform disease status, with studies suggesting a positive correlation between disease burden and HPV ctDNA load.19,20,31–33,46,47 Plasma HPV DNA levels have been found to negatively correlate with overall survival in patients with recurrent, metastatic HPV-positive HNSCC.31 As for HPV ctDNA in other bodily fluids, HPV ctDNA is enriched in the saliva of patients with HPV-positive OPSCC, with higher copies of ctDNA in patients with recurrent locoregional disease compared with patients with only distant disease.48,49 Salivary HPV ctDNA levels in these patients corresponded to treatment response and failure, whereas only plasma HPV ctDNA was associated with worse prognostic scores in paired plasma and salivary samples.49 Collectively, salivary and plasma HPV ctDNA testing can provide complementary information about disease status in patients with HPV-positive OPSCC.
HPV ctDNA may aid in the prognostication of patients with HNSCC, and detectable post-treatment HPV ctDNA can inform clinical outcomes. A study by Dahlstrom et al. did not identify any statistically significant difference in 3-year progression-free survival in patients with HPV-positive OPSCC with and without detectable pretreatment serum HPV DNA (p=0.80); pretreatment serum HPV DNA was also not useful as a marker for disease recurrence (p=0.173).20 Conversely, evidence indicates that post-treatment HPV ctDNA can offer insight into prognosis. Ahn et al. evaluated post-treatment HPV ctDNA with a combination of saliva and plasma samples.22 Assessing HPV ctDNA by qPCR showed 90.7% specificity and 69.5% sensitivity in predicting disease recurrence within 3 years.22 Additionally, a prespecified analysis of post-treatment plasma HPV ctDNA in the NRG-HN002 trial showed an NPV of post-treatment undetectability of 93% and 95% for 2-year locoregional failure and progression-free survival, respectively, in HPV-positive OPSCC.47
Of growing interest is the clinical significance of ctDNA kinetics. An early increase in plasma HPV ctDNA in patients with locally advanced OPSCC receiving chemoradiation (n=34) was associated with superior freedom from progression (p<0.05) and correlated with imaging findings in the primary tumour.50 Chera et al. examined plasma HPV ctDNA clearance in the post-treatment setting and found that, in patients with non-metastatic HNSCC and an initially positive post-treatment test for HPV ctDNA, all patients with disease recurrence (n=15) had persistently elevated HPV ctDNA on subsequent collection, whereas 89% of patients without disease recurrence (n=9) had clearance of their HPV ctDNA.29 Similarly, Fakhry et al. observed that persistent post-treatment detection of oral HPV ctDNA was associated with inferior outcomes with increased risk of recurrence (hazard ratio 3.72; p<0.001) and death (hazard ratio 6.61; p=0.003).21 Furthermore, there is evidence that dynamic changes in HPV ctDNA levels in patients with recurrent or advanced HPV-positive HNSCC can occur before radiographic evidence of response, suggesting a role for HPV ctDNA kinetics in assessing early treatment efficacy.31,34
Studies demonstrate that the incidence, as well as the rate, of HPV ctDNA clearance predicts treatment response in patients with HNSCC. Chera et al. evaluated HPV ctDNA clearance profiles in patients with non-metastatic HPV-positive OPSCC treated with definitive chemoradiation (n=67).32 The authors showed that, in patients with rapid HPV ctDNA clearance, defined as elevated baseline plasma HPV ctDNA levels (>200 copies/mL) and >95% clearance by day 28 of chemoradiation, regional disease-free survival was 100% at 18 months compared with 65% and 90% in patients with unfavourable clearance profiles with and without adverse clinical risk factors (>10 tobacco pack years or T4 disease) (p=0.0049), respectively.32 Similarly, rapid clearance of HPV ctDNA (<1 copy/mL) by postoperative day 1 was observed in patients without pathological risk factors for recurrence after curative intent surgery, whereas HPV ctDNA remained elevated (>350 copies/mL) on postoperative day 1 in patients with risk factors for residual disease (gross extranodal extension, positive margins, distant metastases).51 An exploratory analysis of detectable postoperative HPV ctDNA was associated with clinical risk factors for recurrence, including lymphovascular invasion and extranodal extension, as well as worse recurrence-free survival.52
Cost analysis
Adapting ctDNA testing into clinical practice confers an advantage over tissue biopsy and imaging, as ctDNA assessments are minimally invasive, allow for serial sampling over time and can potentially alleviate the costs of care. Cost modelling by Siravegna et al. estimated costs to be 37% lower when using HPV ctDNA analysis as the diagnostic of choice compared with standard invasive diagnostic approaches requiring tissue biopsies.24
Cost scenarios demonstrate cost savings in surveillance as well as diagnosis with HPV ctDNA. A cross-sectional cost comparison study by Kowalchuk et al. on the post-treatment surveillance of HPV-positive OPSCCs estimated that the median costs of surveillance were lowest when using regular ctDNA testing, followed by surveillance according to the National Comprehensive Cancer Network guidelines and, lastly, surveillance using an imaging-intensive approach that requires PET-CT scans at 1 year and 2 years.53 Surveillance using ctDNA monitoring is anticipated to save costs when HPV ctDNA testing improves quality of life, decreases imaging use and gets drawn less frequently than in clinical trials.54
Future directions
The standard of care for definitive treatment of locally advanced HNSCC involves concurrent chemoradiation or resection followed by adjuvant radiation with or without chemotherapy.45 Given the superior survival outcomes and cure rates of patients with HPV-positive HNSCC, studies are exploring strategies for treatment deintensification to minimize treatment-related toxicity. The phase II clinical trials MC1273 and NRG Oncology HN002 have shown comparable local control rates and overall survival outcomes with dose-reduced radiation therapy during definitive or adjuvant treatment.55,56 However, adopting a deintensification strategy into clinical practice requires data from long-term follow-up and phase III studies.8 HPV ctDNA could serve to guide clinical decisions about treatment deintensification. A deintensification approach driven by HPV ctDNA testing requires rigorous investigation and validation. Clinical trials are on-going to elucidate whether HPV ctDNA can risk stratify individuals with HPV-positive HNSCC and ascertain which subgroup of low-risk patients will benefit from treatment deintensification.57,58
A treatment strategy driven by HPV ctDNA biomarker data needs to be extensively examined in randomized studies, whether for treatment deintensification or intensification. As mentioned earlier in this review, HPV ctDNA is capable of detecting early recurrence following definitive therapy, while rapid HPV ctDNA clearance predicts therapeutic response and risk for recurrence.28,29,30,50,59 However, there are no clear data at this time to inform clinical decisions about initiating or altering therapy in the setting of early recurrence or molecular residual disease. One can envision surveillance using HPV ctDNA to recognize early recurrence and opportunities for salvage therapy, thereby minimizing the morbidity and mortality of disease recurrence or metastasis.58 Clinical trials are necessary to determine whether commencing early treatment or changing systemic therapy for early recurrence or molecular residual disease affects clinical outcomes and whether using HPV ctDNA can reliably individualize treatment plans.18,59
Conclusions
ctDNA is a novel technology with the potential for personalizing the clinical care of patients with HPV-associated HNSCCs. Opportunities to use HPV ctDNA in patients with HPV-positive HNSCC encompass all aspects of care, including screening, diagnosis and surveillance. Accurate HPV ctDNA detection is crucial for applying ctDNA analysis to clinical practice. HPV ctDNA identifies recurrence and early clearance informs treatment response and risk for disease recurrence. Clinical studies are imperative to ascertaining the utility of HPV ctDNA in HPV-positive HNSCC and guiding treatment strategies that adapt to cancer biomarker information from HPV ctDNA.