Abstract
Advances in our understanding of the molecular basis of cancer have shown that biomarkers can help guide therapy in a growing number of cancer types and that targeted therapies are becoming increasingly integral to cancer management. However, how to interpret the rapidly expanding wealth of data on cancer genomics and ensure that the best evidence is effectively translated into an optimal individualised therapeutic strategy, remains a challenge. Caris Target Now™ is a well-established, evidence-based molecular profiling service provided by Caris Life Sciences (Basel, Switzerland). Caris Target Now utilises the latest diagnostic molecular assay technologies to determine the unique biomarkers of a patient’s tumour. This is combined with an extensive review of the published clinical evidence to correlate predictive biomarkers with drug response, in order to reveal the potential benefit (or lack of benefit) of specific therapeutic agents. By this process, Target Now provides tumour-specific information that can be used to help physicians personalise cancer therapy for their patients.
Personalised cancer care is becoming an increasingly important aspect of oncological practice. With the advent of high-throughput molecular profiling technology, it is now possible to identify the unique characteristics of a specific tumour and use this information to tailor treatment to the individual patient. Identification of relevant biomarkers, which include genes, proteins and other molecules, can help to ensure that specific treatments are targeted to those patients who are most likely to gain optimal benefit while avoiding the side effects of ineffective therapy. On a societal level, a personalised approach offers the potential to substantially reduce health care costs, by reducing treatment use in patients unlikely to benefit. This is especially important given the rapidly escalating economic burden of cancer that is, in part, driven by the expense of innovative new treatments.1
Most cancer therapies gain approval despite typical response rates of around only one-third of patients.2 Thus, a significant proportion of cancer patients receive treatment of limited clinical benefit. Better targeted clinical decision-making early in the disease course would improve outcomes for patients. More comprehensive and earlier use of molecular profiling could result in more patients receiving effective therapy at a stage where benefits may be optimal rather than after one or more lines of failed treatment, an especially important consideration in aggressive cancers for which the time window for effective intervention may be limited. It may also mean that patients with rare cancers, which have a limited evidence-base and few treatment options, may be able to benefit from existing targeted therapies. A more personalised approach to patient care, with targeted treatment directed by the molecular profile of the tumour rather than a ‘one-size-fits-all’ strategy, would also lead to more effective allocation of limited financial resources.
Over the past decade, an increasing number of targeted therapies to treat different types of cancer have been introduced and have contributed to improvements in survival rates. Among the first successful targeted agents in cancer were trastuzumab (Herceptin®) for breast cancer overexpressing the Human epidermal growth factor receptor 2 (HER2) receptor and imatinib mesylate (Gleevec®) for abnormal protein tyrosine kinase activity in chronic myeloid leukemia and gastrointestinal stromal tumours. Other examples of targeted therapies include erlotinib (Tarceva®) and gefitinib (Iressa®), both of which target epidermal growth factor receptor (EGFR)-expressing non-small-cell lung cancer (NSCLC), cetuximab (Erbitux®) for patients with EGFR-expressing metastatic colorectal cancer, crizotinib (Xalkori®) for patients with locally advanced or metastatic NSCLC that is anaplastic lymphoma kinase (ALK)-positive, and vemurafenib (Zelboraf®) for BRAF V600E mutation-positive metastatic melanoma. These treatments, and many other targeted therapies, are changing the therapeutic landscape in oncology. However, despite the significant improvements in outcomes seen with targeted therapies, response rates can remain low. For instance, despite its known relationship with HER2 overexpression, trastuzumab yields a response rate of only 35−40 % in patients with HER2-positive breast cancer. It is increasingly evident that focusing on a single molecular target in isolation may not be sufficient, since there is a complex series of inter-relationships between biomarkers, with differential expression of proteins having both an upstream and downstream effect on signal transduction pathways. Thus, a comprehensive knowledge of the relationships between different biomarkers is often required in order to select the most appropriate treatment option.
The need for greater understanding of how biomarkers interact is illustrated by the response seen to the EGFR inhibitors, cetuximab and panitumumab (Vectibix®). These agents bind to EGFR with high affinity, inhibiting tumour proliferation and inducing apoptosis by preventing the activation of downstream signalling pathways (e.g. PI3K/Akt, RAS/Erk and STAT). However, approximately 40 % of patients with metastatic colorectal cancer have a KRAS mutation that can result in constitutive activation of pathways downstream of EGFR, thereby conferring resistance to EGFR inhibitor therapy.3 Because of this, the therapeutic efficacy of EGFR inhibitors is dependent on the presence of wild-type KRAS.4,5 However, it has also been shown that even among patients with wild-type KRAS, less than half respond to EGFR inhibitor therapy.6 This resistance has been attributed to the differential expression of other biomarkers downstream of KRAS (see Figure 1). One study showed that around 10 % of patients with wild-type KRAS had BRAF mutations and were non-responsive to EGFR inhibitor treatment.7 Conversely, all KRAS wild-type patients who did respond had BRAF wild-type. Other EGFR downstream pathways may also be important in conferring EGFR inhibitor resistance. PIK3CA activating mutations, seen in 3−8 % of KRAS wild-type patients, may impair therapeutic response to EGFR inhibition,8,9 as may lack of expression of the PTEN tumour suppressor gene.9,10 The identification of additional genetic determinants of primary resistance to EGFR-targeted therapy demonstrates how a more comprehensive approach to biomarker profiling can improve targeting of therapy. In this case, if all KRAS wild-type tumours were also evaluated for BRAF, PIK3CA and PTEN status, more non-responders to EGFR inhibitor therapy could be excluded from treatment (see Figure 2).
As our knowledge of biomarkers increases, it is also becoming evident that many important molecular targets, typically associated with one or two tumour types, are actually expressed in cancers of multiple lineages. For instance, HER2 overexpression occurs in 25–30 % of breast cancers and up to 25 % of gastric cancers, indications for which trastuzumab has been approved. However, 34 % of gastroesophageal cancers, 11−30 % of ovarian cancers and 11 % of colorectal cancers also overexpress HER2.11 Thus, a patient with colorectal cancer overexpressing HER2 could theoretically benefit from HER2 targeted treatment if other standard of care options have failed. Similarly, low levels of O6-methylguanine-DNA methyltransferase (MGMT) expression are associated with response to temozolomide (Temodar®/Temodal®) in patients with glioblastoma multiforme, a type of brain cancer. MGMT is an enzyme that repairs methylated DNA. Temozolomide works by triggering DNA methylation in tumour cells, which causes cells to enter apoptosis. Because MGMT repairs the DNA methylation caused by temozolomide, overexpression of MGMT can mean temozolomide is less effective at disrupting tumour growth. Low levels of MGMT expression are also seen in patients with several other cancer types, including melanoma, gastroesophageal cancer, NSCLC and breast cancer. Thus, it has been postulated that treatment with temozolomide might be of benefit in tumour types other than brain cancer. As an example of this, Shacham-Shmeuli et al. reported the use of temozolomide in two patients with metastatic colorectal cancer with loss of MGMT expression, with progression-free survival (PFS) of five and six months being achieved despite active disease progression during their previous two lines of therapy.12 However, it is unlikely that large-scale randomised controlled trials to support targeted therapy outside of initially approved indications will be conducted, due to the difficulty in recruiting sufficient numbers of patients, as well as the often limited patent protection after lengthy secondary development.Advances in our understanding of the molecular basis of cancer have shown that biomarkers can help guide therapy in a growing number of cancer types and targeted therapeutics are becoming increasingly integral to cancer management. However, how to interpret the rapidly expanding wealth of data on cancer genomics and ensure that the best evidence is effectively translated into an optimal individualised therapeutic strategy remains a challenge. One potential approach that is gaining momentum is the use of evidence-driven molecular profiling services to guide targeted therapy choices.
Target Now™
Caris Target Now is a well-established evidence-based molecular profiling service provided by Caris Life Sciences (Basel, Switzerland). Caris Target Now utilises the latest diagnostic molecular assay technologies to determine the unique biomarkers of a patient’s tumour. This is combined with an extensive review of the clinical evidence base to correlate predictive biomarkers with drug response, in order to reveal the benefit (or lack of benefit) of specific therapeutic agents. By this process, Target Now provides tumour-specific information that can be used to help physicians personalise cancer therapy for their patients.
Target Now is suitable for any patient with a solid tumour diagnosis. The stage in the clinical pathway where the analysis is performed will depend on the individual patient. However, it is most frequently used for patients with advanced disease receiving standard of care treatment where multiple options are available but the optimal therapeutic strategy is unclear, and patients for whom standard of care options have been exhausted or are not an option, for example due to co-morbidities. In addition, patients with highly aggressive or rare tumours, advanced disease or a good performance status and a wish to continue with further treatment options may also be appropriate candidates.
Target Now uses the latest technologies to provide a detailed molecular profile and biomarker status of tumour tissue. Immunohistochemistry (IHC) determines the level of expression of protein biomarkers. Gene deletions, amplifications, translocations and fusions are detected through fluorescent or chromogenic in situ hybridisation (FISH/CISH). Reverse-transcriptase polymerase chain reaction (RT-PCR) is used to measure RNA expression of several genes, including Topoisomerase 2 (Topo-2), Class III B-Tubulin (TUBB3), Thymidylate synthase (TS) and breast cancer susceptibility gene (BRCA). Direct gene sequencing is used to identify mutations in targeted genes, including EGFR, PI3KCA, KRAS, BRAF, (tyrosine-protein kinase Kit) CKIT and neuroblastoma RAS viral oncogene homolog (NRAS). The biomarker analysis applies the technology as best described in the clinical literature and is based on the most advanced, state-of-the-art biomarker tests validated to the highest regulatory quality standards. This approach ensures that the best clinical evidence is applied to the individual patient. Target Now is technology independent and so its inherent flexibility allows the rapid adaptation of innovative and validated technology platforms as they emerge from the research environment and are incorporated into routine practice. Given the acceleration in development of personalised medicine, this incorporation of new technologies is necessary to maintain a relevant and actionable report for the treating clinician.It is important that the biomarker tests follow the test protocols and technologies as described in the literature. It is not clinically reasonable to expect changes in gene expression to be a valid surrogate for differential biomarker expression, if the supporting evidence describes changes in protein expression. For example, exploratory biomarker studies assessing EGFR protein expression using IHC reported mixed results on the link between EGFR expression and outcome with gefitinib, with no consistent or conclusive relationship identified.13,14 Initial retrospective analysis in a small number of patients showed an association between high EGFR gene copy number and gefitinib treatment outcomes15 but this could not be confirmed in a larger trial. The strongest evidence has come from the discovery of sensitising mutations of EGFR in patients with advanced NSCLC, which was confirmed in large scale, prospective, randomised controlled studies.16 This does not mean that EGFR results from other assay platforms should be disregarded, as recent data has linked the EGFR H-score as measured by IHC to a higher likelihood of benefit with cetuximab treatment in NSCLC.17 Target Now™ incorporates both evidence-based technologies with results linked to different chemotherapeutic agents so that the final report can be as informative as possible to the treating physician.
All assay platforms utilised in Target Now are established and use approved commercially available tests and materials where available. Any laboratory developed tests are validated according to Clinical Laboratory Improvement Amendments (CLIA)/New York State laboratory license requirements. No novel or proprietary tests are performed.
At present, all tests are performed at the central Caris CLIA-certified laboratory in Phoenix, AZ, USA, although European laboratories are being considered. Tissue can be sourced from recently obtained or archived biopsy or resection tumour specimens that are formalin-fixed and paraffin embedded. Recently obtained specimens are strongly preferred as the biomarker profile of a tumour is known to alter over time and with treatment.18-20 high throughput of patient tumour samples means that practical experience with newer tests and technologies occurs rapidly. Strong laboratory technical expertise and continual investment in the best pathology and laboratory molecular genetics are a crucial part of the Target now molecular profiling service.
Pathology and molecular genetics play a critical role in Target now. Board-certified, experienced oncologic pathologists oversee molecular testing from the time the test is ordered to the generation of a final Target now report.
The Target now oncologic Pathology team works with skilled molecular laboratory staff to maintain the highest quality and integrity of Target now laboratory data and data analysis. The pathologists review the submitted blocks or slides, confirm the diagnosis, and evaluate the volume of tumour available for various molecular studies. Potential discrepancies are carefully assessed by appropriate confirmatory studies and the Target now pathologist may communicate directly with the treating oncologist to discuss prioritisation of biomarker assays when there is insufficient biopsy material to complete the full panel of tests ordered. In this situation, the goal is to maximise clinical results for the patient by selecting relevant biomarkers with the highest clinical impact.
The results of IHC stains and CISH assays are read by caris molecular pathologists who also perform quality control of laboratory results on various technology platforms to ensure the correct and appropriate association of clinical benefit or lack of benefit for each biomarker test result before the final report is signed.critically, the comprehensive biomarker analysis is combined with extensive assessment and review of the clinical literature to ensure optimal treatment associations are made based on the patient’s individual tumour profile. over 100,000 clinical studies have been reviewed, of which around 6500 to date have been included in a regularly updated database. An evidence team, consisting of molecular biologists, oncologists and pathologists, review and evaluate biomarker studies based on uS Preventative Services Task Force (uSPTF) level of evidence criteria. once studies have been graded, an evidence review board reviews each biomarker and decides whether the evidence is sufficiently robust for inclusion in the evidence database. This evidence is used as the basis for treatment suggestions based on a personal biomarker analysis. These suggestions include treatments both likely and unlikely to be of benefit on the basis of the biomarker profile. In many patients, previously unconsidered treatment options may be revealed and ineffective treatments can be avoided.
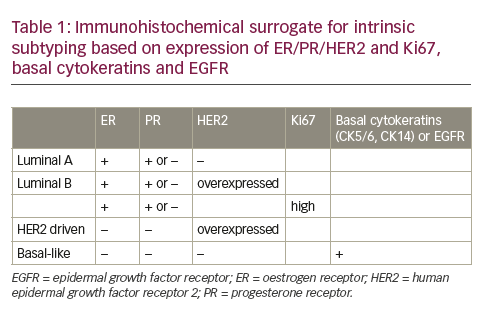
Two distinct profiling options are offered as part of Target now (see Table 1). Target now Select™ provides a focused biomarker analysis for standard, tumour-specific therapeutic associations for five major tumour types (breast, colorectal, nSclc, ovarian surface epithelial and melanoma). Analysing approximately 10–20 cancer-specific biomarkers, Target now Select offers rapid turnaround times of 7−9 days and efficient use of smaller samples (e.g. fine needle aspiration samples). Target now comprehensive™ offers a broader panel of biomarker tests for therapeutic associations across all tumour types, including approximately 30–35 biomarkers. caris Target now comprehensive has six independent profiles available (breast, colorectal, nSclc, ovarian surface epithelial, melanoma, and all other solid tumours). Both Target now Select and comprehensive include on and off national comprehensive cancer network (nccn) compendium drug associations, based on the range of relevant evidence.
A fully referenced report detailing the molecular profile together with treatment options likely and unlikely to be associated with potential benefit is produced. For each treatment option, correlated biomarker analysis methods and results are provided together with an indication of the overall strength of evidence (high, intermediate or low) and key references on which this evidence is based. The report, which can be made available online, is provided within 15 days, meaning that the clinician has the best available evidence to hand in due time to support treatment decisions for the individual patient.
In the future, Target now will also identify open, relevant clinical trials based on the patient’s expressed biomarkers with clinical Trials connector™
Clinical Evidence
To date, there are just a few studies that show the clinical benefit of this type of molecular profiling to guide therapy. A feasibility study, in which 186 patients had tumour biopsies submitted for IHC and mA, demonstrated that it was already possible to receive and process high-quality tissue specimens on a consistent basis in 2006.21 This study also illustrated the high proportion of patients in which it was possible to identify a target in patients’ tumours for which there existed a potential therapeutic agent. Based on these promising results, a study in which the treatment decision was based on individual tumour profiling was initiated to investigate whether molecular targeted therapy could result in improved outcomes. The Bisgrove Study
The Bisgrove study was the first clinical trial to assess the use of molecular profiling of tumours to identify treatment targets in patients with refractory cancers.22 In this prospective multicentre trial, patients with metastatic cancer with documented disease progression on their most recent line of systemic therapy had tissue samples submitted for molecular profiling by IHC/FISh and mA. Treatment was then selected based on targets identified by molecular profiling. The primary objective of the study was to compare PFS with a treatment regimen based on molecular profiling with PFS (more correctly defined as time to progression, since death is excluded as an event) observed for the patient’s most recent prior treatment regimen. molecular profiling was considered to be clinically beneficial for patients who had a PFS ratio (on molecular profiling selected therapy versus on prior therapy) of ≥1.3.
Of 86 patients who had molecular profiling attempted, a molecular target was identified in eighty-four. Sixty-six of these patients were treated based on results of molecular profiling (breast cancer, n=18; colorectal, n=11; ovarian, n=5; other, n=32). overall, more than onequarter of treated patients (n=18, 27%) had a PFS ratio ≥1.3. median PFS ratio was 2.9 (1.3–8.15) (see Figure 3). When assessed by tumour type, a PFS ratio ≥1.3 was achieved by 44 % of patients with breast cancer, 36 % with colorectal cancer and 20 % of patients with ovarian cancer. overall survival for the 18 patients with a PFS ratio ≥1.3 was 9.7 months, compared with five months for all 66 patients.
One patient had a complete response and five patients had partial responses. Fourteen patients (21 %) were without progression after four months. Treatments selected based on molecular profiling were generally well tolerated with no treatment-related deaths and only nine treatment-related serious adverse events.
Of interest, there was no concordance between the treatment given based on molecular profiling and the treatment the physician indicated would have been used if molecular profiling data were not available.
Other Studies
Other data also suggest molecular profiling may have a positive effect on treatment outcomes. In a single-centre pilot study of patients with advanced refractory solid tumours, six of nine patients who received treatment guided by molecular profiling (Target Now) showed a clinical response.23 In another pilot study in patients with adenoid cystic carcinoma, four of six patients showed at least a partial response over six months to molecular profile-directed therapy, including one patient who initially had multiple lung, liver and bone metastases with no evidence of disease for over 15 months.24
Similarly, in a retrospective study conducted at the University of Texas MD Anderson Cancer Center, treatment directed against a single molecular aberration in a patient’s tumour resulted in higher response rates and extended PFS compared with non-matched therapy.25 In this study, clinical outcomes were assessed in patients with metastatic melanoma who were tested for tumour molecular aberrations and entered into a Phase I trial with a matched targeted agent (n=77) or who were treated in a Phase I trial without regard for their molecular profile (n=83). Overall response rate was 39 % in patients treated with a matched targeted treatment, compared with 9 % in patients not treated with matched therapy (p=0.0018). Median PFS was also longer on matched versus non-matched therapy (5.3 vs 3.7 months, p=0.004).More evidence should be provided by the international WINTHER trial, which aims to enrol 200 patients with advanced cancer and will compare PFS with last treatment line (standard treatment) before inclusion in the study with PFS with individualised targeted therapy based on molecular profiling. Individualised therapy will include two treatment arms, one in which treatment will be based on known abnormalities of tumour DNA (i.e. matched molecular targeted therapy) and a second arm in which treatment will be based on a predictive drug efficacy score after RNA analysis of tumour and normal tissue to identify relevant aberrations. This means all patients can be included, including the 60−70 % of patients that do not show known genomic aberrations of tumour DNA.
Caris Registry
To provide further evidence of the effectiveness of clinical decision-making based on molecular profiling, Caris Life Sciences has established a registry of patients who have been profiled as part of Target Now™. This registry is a prospective, multicentre, observational outcomes database that will provide information on biomarker expression, treatment and clinical outcomes. Data will be collected at baseline (date of Target Now report) and then every nine months for five years or until death. It is hoped this will provide a valuable resource linking tumour biomarker data with clinical outcomes.
Conclusions
Identifying clinically relevant and patient-specific biomarkers can help guide more effective and targeted treatment. As treatment costs in oncology continue to rise, the need for smarter, more accurate targeting of treatments will become increasingly imperative. As part of this, there is a requirement for evidence-based strategies that help translate the growing body of complex biomarker data into clinically relevant treatment recommendations. Caris Target Now, a molecular profiling service that combines state-of-the-art biomarker analysis with systematic evidence-based literature review to offer a rational, personalised therapeutic strategy, is one such approach with the potential to improve clinical outcomes for appropriate patients.