Carcinoma of the uterine cervix is the second most common form of cancer in women worldwide. In the Czech Republic, it has an incidence of 19/100 000 women and a mortality rate of 7/100,000 women.1 There are two main types of cervical cancer, squamous cell carcinoma and adenocarcinoma, as well as a variety of much less common types (endometroid adenocarcinoma, clear cell adenocarcinoma, serous adenocarcinoma, mesonephris adenocarcinoma, adenosquamous carcinoma, adenoid cystis carcinoma, small cell carcinoma and metastatic tumours). Among the two main types, squamous cell carcinoma (see Figure 1) is more common than adenocarcinoma by a ratio of approximately 85:15.
Several pre-malignant stages, known as cervical intraepithelial neoplasia (CIN) or cervical dysplasia, are related to the development of invasive carcinoma. These stages can be distinguished and graded histologically. Changes in less than one-third of epithelial cells indicate mild dysplasia (CIN I). Defects in cell maturation and nucleocytoplasmic ratio changes with frequent mitosis in two-thirds of epithelial cells are characteristic of moderate-to-marked dysplasia (CIN II). Defects in cell maturation with frequent atypical mitosis across the entire epithelium indicate severe dysplasia/carcinoma in situ (CIN III). The ratio of pre-malignant squamocellular cancer to glandular neoplasia is about 60–80:1. Risk factors for the development of cervical cancer include a high number of sexual partners, multiparity, young age at first pregnancy, smoking, ethnicity, sexual intercourse before the age of 16, immunodeficiency, coincidence of sexually transmitted diseases, and other factors such as vitamin C and folate deficiency.2,3
The genetic basis of progression from CIN I to CIN II/III and on to invasive carcinoma is poorly understood. Although infection with high-risk human papillomavirus (HPV) is recognised as an essential initiating event in cervical tumourigenesis,4 this alone is not sufficient to explain the progression to invasive cancer. A number of cell regulators are involved in the process. Certain chromosomal aberrations have been associated with the progression of CIN to carcinoma, especially the amplification of the human telomerase gene hTERC (3q26) and the myelocytomatosis-C proto-oncogene MYCC (8q24);5,6 hTERC is involved in the maintenance of chromosomes by providing telomerase stability and regulating telomerase length; MYCC is a transcription factor that participates in cell proliferation and differentiation.7
Cytogenetic detection of structural aberrations using banding analysis is often problematic with human solid tumours, because of the difficulty in obtaining high-quality banded metaphase preparations. Fluorescence in situ hybridisation (FISH), developed at the end of the 1980s, is an important method for detecting and characterising chromosomal aberrations in human cancer. It uses the base pairing of marked DNA probes with their complementary target DNA sequences in metaphase chromosomes or interphase cells. FISH with probes for specific genes enables to assess the copy number of these genes on a cell-by-cell basis and thus provides information about the heterogeneity in the tumour. In addition, the detection of these cells when present at low frequency may be prognostically important.
Advances in molecular cytogenetics techniques have enabled the study of genomic alterations as biomarkers of progression during uterine tumourigenesis. This is the basis of the rationale for the development of a specific FISH assay intented as a cytogenetic diagnostic tool for the direct detection of HPV-infected cells and alterations of selected genes in cytology specimens (HPV-FISH). Recently, a new FISH probe kit was designed to identify HPV-infected cells and determine copy number changes for hTERC and MYCC in pre-neoplastic lesions and cervical carcinomas.8,9
In a study conducted by our group at Masaryk University in Brno (Czech Republic), the cytology specimens of 26 women with a histologically confirmed diagnosis of either CIN or cervical cancer were evaluated alongside routinely collected cytology samples (Pap smears) using the new HPV-FISH assay, and the cytogenetic findings were compared with the histological diagnoses. We also set the prognostic risk for each patient according to recommended settings.
Patients and Methods
Cervical Specimens
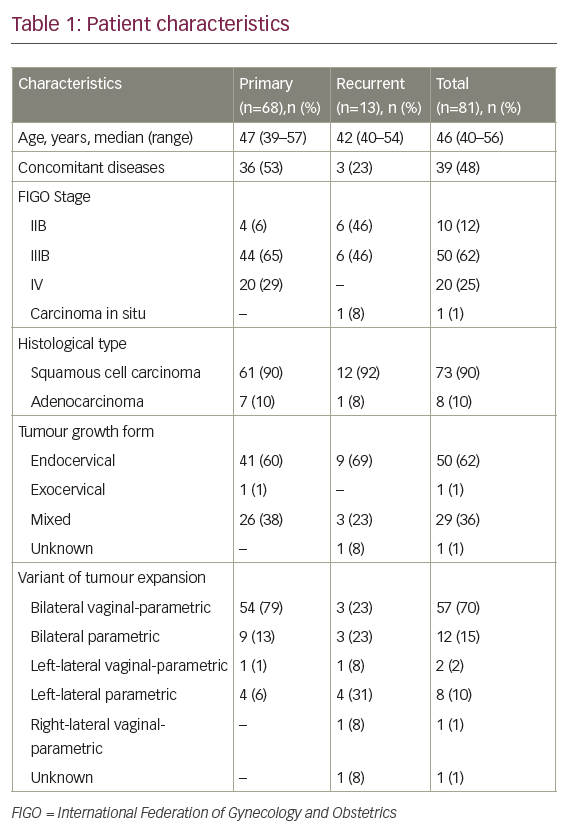
Cervical specimens were obtained from the Department of Gynaecological Oncology at the Masaryk Memorial Cancer Institute in Brno. Twenty-six Pap smears were included: those of nine patients with pre-malignant lesions (median age 40 years; range 24–64 years) and those of 17 patients with a diagnosis of cervical carcinoma (median age 42 years; range 31–69 years). Based on histology classification, there were two cases of CIN I, one case of CIN II and six cases of CIN III among patients with pre-malignant lesions, and two cases of carcinoma in situ, 12 cases of TNM (tumour, node, metastasis) stage IB cancer and three cases with TNM stage IIIB cancer among patients with cervical carcinoma.
Slides from Pap smear tests were prepared according to standard procedures. Slides were then fixed in methanol for 20 minutes at 4 °C and in 3:1 methanol:acetic acid for another 20 minutes at 4 °C. Slides were dried at room temperature and then stored at -20 °C until required for hybridisation.
Assay Description
The Vysis Cervical FISH Probe Kit® (Vysis Inc/Abbott Molecular, Abbott Park, US) was used to identify HPV-infected cells and determine copy number changes in the chromosomal regions 3q26 (hTERC) and 8q24 (MYCC). This kit enables identification of high-risk HPV types (16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66 and 68) using biotin labelling and a tyramide signal amplification (TSA) assay. Detection of copy number alterations of the hTERC and MYCC genes is achieved using standard FISH techniques. The hTERC probe is labelled with a SpectrumGold fluorescent label and covers a region approximately 495 kilobases (kb) in length. The MYCC probe is labelled with a SpectrumRed fluorescent label and covers a region approximately 821 kb in length.
Slide Pre-treatment and Hybridisation
Slide pre-treatment and FISH were performed according to the manufacturer’s protocol. Slides were briefly soaked in 2X saline sodium citrate (SSC) buffer at 73 °C for two minutes and then incubated in pepsin (0.5 mg/ml in 10 mmol/l hydrochloric acid) at 37 °C for 10 minutes. They were than soaked in 1X phosphate buffered saline (PBS) at room temperature for five minutes, fixed in 1 % neutral-buffered formalin at room temperature for five minutes, and finally soaked in 1X PBS at room temperature for another five minutes.
Slides were dehydrated using 70 %, then 80 % and 96 % ethanol for one minute before air drying. The probe mixture (10 μl) was then applied and the slides and probe were co-denatured at 72 °C for two minutes and hybridised at 37 °C overnight. After hybridisation, the slides were washed in 2X SSC at 48 °C for two minutes and then in 2X SSC at room temperature for one minute.
Detection of Human Papillomavirus Using Tyramide Signal Amplification Assay
Detection of the biotin-labelled HPV probes was performed using an Alexa Fluor® 488 TSA kit (Invitrogen/Life Technologies, Carlsbad, US) according to the directions given in the Vysis Cervical FISH Probe Kit. The slides were incubated in 3 % hydrogen peroxide at room temperature for 30 minutes to block endogenous peroxidase activity and then soaked in 1X PBS at room temperature for five minutes. Prior to application of streptavidin-HRP (SA-HRP) conjugate (diluted 1:100 in blocking reagent), the slides were incubated with 1 % blocking reagent in PBS (both incubations were performed in a humified chamber at 37 °C for 25 minutes). After washing the slides three times in 1X PBS at 37 °C for five minutes, the biotin-labelled HPV probe-SA-HRP complex was visualised by incubation with Alexa Fluor 488 labelled tyramide (1:100 dilution) for 10 minutes at room temperature. The slides were then washed three times in 1X PBS at 37 °C for five minutes and diamidino-2-phenylindole (DAPI) nuclear counterstain was applied.
Slide Analysis
The slides were analysed under an Olympus BX-61® fluorescence microscope equipped with a CCD-1300D® Vosskühler camera and LUCIA-KARYO/FISH/CGH® imaging system (Laboratory Imaging, Czech Republic) using 40x and 100x magnification and DAPI with green, gold and red single bandpass filter sets. In all cases, the entire hybridised surface area was analysed. Cells were evaluated according to the Vysis Cervical FISH Probe Kit directions.
Classification Algorithm
A green filter was used to visualise HPV staining, its localisation in the nucleus was confirmed by DAPI staining. The staining pattern was classified as diffuse or punctate (see Figure 2), as described in previous studies.8,9 The sample was considered positive for HPV infection if at least one HPV-positive cell was found. All HPV-positive cells and their staining patterns were recorded. The number of hTERC and MYCC signals was then determined for each HPV-positive cell. The patient was considered positive for chromosomal aberration if four or more HPV-positive cells demonstrated copy number gain (more than two fluorescent signals) at at least one chromosome locus (hTERC or MYCC). Otherwise, the patient was considered to be HPV-positive but chromosome-negative.
In the absence of HPV-positive cells, the slide was evaluated for the presence of cells with amplified hTERC and/or MYCC. The patient was considered positive for chromosomal aberrations if >5.8 % of cells (the set cut-off level from negative controls) demonstrated copy number gain (more than two fluorescent signals) at at least one chromosome locus (hTERC or MYCC). Otherwise, the patient was considered HPV- and chromosome-negative.
Patients were classified as low-risk, medium-risk or high-risk according to the HPV and chromosomal findings (see Figure 3).
Results
The results regarding the numbers of patients with HPV-positive cells and chromosomal abnormalities, and the risk assessment for patients with CIN or cervical cancer are summarised in Tables 1 and 2.
HIV-positive cells were found in 22 of 26 patients (85 %). Three of nine patients with pre-malignant lesions (CIN I, CIN II and CIN III) were found to be HPV-negative; only one patient (with carcinoma in situ) of the 17 with cervical cancer was found to be HPV-negative.
The mean number of HPV-infected cells in the nine patients with CIN was 61 in the CIN I category (range 0–121; all cases with punctate pattern) and 33 in the CIN III category (range 0–79; three cases with punctate pattern and two cases with punctate and diffuse pattern); no patients in the CIN II category had HPV-infected cells. Among the 17 patients with tumours, the mean number of HPV-positive cells was 85 in the two patients with carcinoma in situ (range 0–170; both cases with punctate and diffuse pattern); 244 in the 12 patients with grade IB carcinoma (range 5–1,974; eight cases with punctate pattern and four cases with punctate and diffuse pattern); and seven in the three patients with grade IIIB carcinoma (range 3–15; all cases with punctate pattern).
Amplification of MYCC (8q24) was found in 11 of 26 patients (42 %) and copy number changes of the hTERC locus in 16 of 26 patients (62 %).
Among the nine patients with CIN (see Table 1), MYCC and hTERC copy number changes were found in three patients (44 %), all with CIN III histology and all categorised as high-risk. Low-risk patients were found only at CIN I (one case) and CIN II (one case) stages.
Among the 17 patients with cervical cancer (see Table 2), hTERC copy number changes were found in 13 individuals (76 %); hTERC alterations were detected in one of the two patients with carcinoma in situ, nine of the 12 patients with TNM stage IB cancer, and all three patients with TNM stage IIIB cancer. Copy number changes for MYCC (8q24) were found in eight patients (47 %) – one with carcinoma in situ, six with TNM stage IB cancer and one with TNM stage IIIB cancer. Amplification of hTERC was detected in all three patients with TNM stage IIIB tumours; however, no copy number changes were found in three of the 12 patients with TNM stage IB tumours. No single amplification of MYCC gene was detected.
All patients with stage IIIB tumours as well as nine of the 12 patients with stage IB tumours were categorised as high-risk. The remaining three stage IB cases were classified as medium-risk and one patient with carcinoma in situ was put in the low-risk category.
Six patients with cervical cancer were positive for lymphovascular space invasion (LVSI). Three of them were stage IIIB patients and the other three were stage IB patients. All six had amplification of hTERC and all were put in the high-risk group.
Discussion
During the last decades, the aetiological role of HPV infection in the development of cervical dysplasia and cervical carcinoma has been well documented. HPV is one of the most common sexually transmitted diseases. The rate of transmission depends on host susceptibility, duration of contact and dose of HPV. The virus enters the organism through microscopic wounds or via direct contact in the cervical transformation zone and infects the basal layer of the epithelium. It is occasionally transmitted from mother to baby during a vaginal delivery.
Infection of cervical epithelial cells with high-risk HPV represents a necessary factor in the development of cervical cancer. More than 15 types of high-risk HPV are now considered oncogenic, HPV 16 being the most dangerous.10 Oncogenic forms of HPV have been found in about 99 % of cervical carcinomas,4,11 and the use of HPV detection with Pap smear examination has increased sensitivity for the detection of dysplasia to almost 100 %.12 However, HPV screening itself is not sufficient to predict which CIN lesions will become malignant, because only a fraction of HPV-infected women progress from mild dysplasia to high-grade lesions and cancer.13 This denotes a possible role for other factors in cervical tumourogenesis.
Genetic abnormalities play an important role in the aetiology of tumour transformation. Chromosomal instability, either numeric or structural, is a characteristic marker of malignant tumours, and has prognostic and predictive impact in many cancers. Specific chromosomal changes have repeatedly been found, mainly in the 3q26 region of hTERC14 and in the 8q24 region of MYCC,5 in malignant and pre-malignant lesions in the cervix and in cervical carcinoma cell lines. According to published data, 80–90 % of cervical carcinoma cases are characterised by amplification of hTERC.15 This specific genetic abnormality was also found in pre-malignant CIN II/CIN III and is thought to be a genetic aberration that occurs in the early stages of tumour development. As such, it has value for predicting malignant transformation and disease progression.14,16,17
Another specific genetic abnormality linked to the development of cervical carcinoma is MYCC amplification. Golijow et al.5 showed that there is a difference in the number of MYCC copies in CIN I, CIN II/CIN III and carcinoma in situ, suggesting that the amplification of MYCC is important not only in tumour progression, but also in cell transformation during pre-invasive stages.
The principal aim of our study was to optimise the HPV-FISH assay (originally developed by Sokolova et al.9) and evaluate if it could serve as a diagnostic test to confirm cytopathological or histopathological findings from cervical lesions. The Vysis Cervical FISH Probe Kit contains triple-colour probes for hTERC (3q26) and MYCC (8q24), together with a probe ‘cocktail’ of six HPV types with homology with the majority of high-risk types. The use of fluorescent probes to detect aneusomy in genomic regions that are sensitive to destabilisation may be an essential tool for identifying cells at risk of progression from mild and moderate to severe dysplasia.
In our set of patients, HPV-positive cells were found in 22 of 26 cases (85 %). Previous studies using HPV-FISH assays showed a 91 % concordance between in situ HPV and polymerase chain reaction (PCR) for the detection of high-risk HPV.8 There was no control PCR-based HPV detection in this study; the presence of HPV-positive cells was established using only the HPV-FISH assay and was confirmed by a pathologist. Unlike PCR, the HPV-FISH assay is able to determine the physical status of the virus (that is, episomal or integrated) and the percentage of infected cells in a sample. Homogenous (diffuse) staining is suggestive of episomal HPV and punctate staining is suggestive of integrated HPV. Algeciras-Schimnich and colleagues showed that the number of cells with punctate staining in pre-cancerous lesions increased with increased cytology classification.8 In our study, we found diffuse staining (episomal HPV) only in one CIN III case and in some carcinoma cases; however, our group of patients was too small to draw any conclusions about this.
In our study, the incidence of hTERC amplification was higher than that of MYCC amplification (62 % and 42 %, respectively). In patients with pre-malignant lesions, MYCC and hTERC copy number changes were found in three of nine cases (44 %), all of whom had CIN III histology. In patients with tumours, hTERC amplification was found in 13 of 17 cases (76 %) and MYCC amplification in eight cases (47 %). An increase of 3q24–28 has been shown to be the most frequent chromosomal aberration in cervical cancer: more than 85 % of invasive cervical carcinoma feature this change.6 Other studies have shown that an increase in the 3q chromosome arm is the most common aberration, not only in invasive carcinoma, but also in CIN II, CIN III and carcinoma in situ.17–20 Using FISH, amplification of hTERC was detected in only some cytologically normal or CIN I cases, but in 62.5 % of CIN II and 76.4 % of CIN III cases.14 No pre-malignant lesion with subsequent regression had hTERC amplification. Subsequent studies have confirmed that increases in hTERC may predict progression from early pre-invasive stages (CIN I/CIN II) to a later stage (CIN II) and to invasive cervical carcinoma.6,16,17 In accordance with these studies, we only found amplification of hTERC in CIN III pre-malignant cases and in the majority of invasive cancers. Our results confirm that the vast majority of invasive cervical carcinomas carry extra copies of hTERC.
In our study, amplification of hTERC in all three CIN III pre-malignant cases was attended by MYCC amplification. Golijow et al.5 have shown significant differences in the degree of MYCC amplification between cytologically normal and abnormal pre-cancerous specimens. Their study also showed higher numbers of MYCC with increased histological classification and confirmed the important role of this gene in pre-malignant cell transformation.
We found a higher incidence of MYCC increase in patients with tumours – eight of them (47 %) had MYCC alterations. Using immunohistochemical techniques, Brenna et al.21 detected a similar incidence of MYCC expression (40 %) in a group of 220 women with cervical carcinoma; 19 % of women with stage I, 33 % of women with stage II and 48 % women with stage III cervical carcinoma according to the International Federation of Gynecology and Obstetrics (FIGO) classification were positive for MYCC; moreover, overall survival was shorter in patients who were positive for MYCC. In our study, MYCC changes were also noted predominantly in patients with stage IB and IIIB tumours, supporting the theory of a role for MYCC in the later stages of cervical tumourogenesis.
By scoring the HPV staining and chromosomal changes, we were able to divide patients into high-risk, medium-risk and low-risk groups. These groups may predict the risk of progression or regression, especially in the pre-malignancy stages. Patients with CIN I and CIN II were defined as low- or medium-risk. Only three patients with CIN III histology were classified in the high-risk group; all of them had both hTERC and MYCC amplification.
Among the 17 patients with cervical cancer, only one (with carcinoma in situ) was classified as being low-risk, because of the absence of HPV-infected cells. Three of the 12 patients with stage IB cancer were defined as being medium-risk. The other nine patients with stage IB cancer, as well as the other patient with carcinoma in situ and all three patients with stage IIIB tumours were categorised as high-risk.
Due to a short clinical follow-up, no statistical analysis of cervical cancer progression was conducted for patients with aberrations (we plan to prospectively follow-up participants for five years after diagnosis). However, six patients were positive for LVSI; all of them were high-risk and had hTERC amplification, and two also had MYCC amplification. Based on these results, we suggest that high-risk patients would require more frequent follow-up and aggressive therapy.
Conclusions
Recent advances in molecular genetics and cytogenetics have enabled new techniques and biomarkers, in particular those used to predict response to therapy or disease prognosis, to become part of routine clinical practice. The preliminary results of our study showed that the HPV-FISH assay is a reliable cytogenetic test for use in clinical practice in order to determine which cervical samples have high-risk HPV infection and chromosomal aberrations with potential to progress to later pre-invasive stages and hence to invasive cervical carcinoma. We also found that the detection of hTERC and MYCC amplifications by FISH improves the risk assessment of patients with cervical carcinoma. It should, however, be borne in mind that the HPV-FISH assay was tested in a small number of patients over a short monitoring period. More detailed analyses of larger numbers of patients and longer follow-up periods are required to confirm the prognostic value of this test. ■