Anaplastic Lymphoma Kinase
Anaplastic Lymphoma Kinase
Located at 2p23, the anaplastic lymphoma kinase (ALK) gene codes for a receptor tyrosine kinase (RTK). This gene was initially identified in the chimeric nucleophosmin (NPM)-ALK fusion driven by the t(2;5)(p23;q35), a recurrent translocation in anaplastic large cell lymphoma (ALCL).1 The ALK gene contains 6,226bp and encodes a 177kDa protein that is transformed to a mature ALK RTK of approximately 200kDa by posttranslational modifications such as N-glycosylation.2 In common with other RTKs, ALK consists of a large extracellular ligand-binding domain, a lipophilic transmembrane-spanning segment and a cytoplasmic tyrosine kinase catalytic region (see Figure 1). Due to the significant homology of the extracellular domain to leukocyte tyrosine kinase (LTK), ALK is classified in the insulin-receptor superfamily of RTKs; the function of the ALK receptor is still poorly characterised. Expression of ALK in normal tissues is restricted to rare scattered neural cells, pericytes and endothelial cells in the brain.1–3 Recent data suggest that ALK plays a role in embryonic neural development and differentiation.4,5
Anaplastic Lymphoma Kinase Rearrangements in Lymphoma
Anaplastic Lymphoma Kinase-positive Anaplastic Large Cell Lymphoma
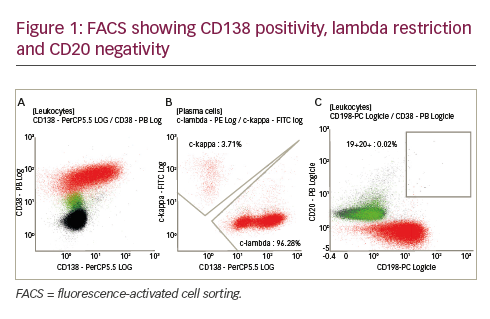
ALK+ ALCL (informally ALKoma) accounts for approximately 60–80% of all ALCL cases.4,6,7 In the World Health Organization (WHO) classification of haematolymphoid neoplasms, ALK+ ALCL is recognised as a separate entity among mature T-cell lymphomas.8 These tumours form a homogenous group in terms of the immunophenotype and clinical behaviour, but demonstrate a heterogeneous morphology. The common type, composed of atypical large, pleomorphic cells with an abundant cytoplasm, is seen in approximately 80% of cases, and morphological variants, including lymphohistiocytic, small- and giant-cell types, are observed in the remaining cases.9,10 ALK+ ALCL cases exhibit a cytotoxic T-cell or null (i.e. lacking expression of both T- and B-cell markers) phenotype and frequently express epithelial membrane antigen (EMA) and CD30. The disease tends to occur in the first three decades of life, with mean age at onset <30 years, and shows a slight male predominance (male to female [M:F] ratio of 1.5–1).4,6–18 Although ALK+ ALCLs are highly aggressive, patients with this lymphoma have a more favourable prognosis compared with ALK-negative ALCL (ALK- ALCL) patients; the overall five-year survival rate approaches 80% in ALK+ ALCL and 48% in ALK- ALCL.4,8,15,17 Multivariate analysis has shown that good prognosis of patients with ALK+ ALCL strongly correlates with expression of ALK, but not with younger age and/or presence in low international prognostic index risk groups.4,15 The inappropriate expression of ALK in ALK+ ALCL derives from chromosomal translocations targeting the 2p23/ALK locus. Occurring in 84% of ALK+ ALCL, the most frequent, t(2;5)(p23;q35), results in the fusion of the amino-terminal end of NPM (currently designated as NPM1) with the intracytoplasmic portion of ALK.1,8,19,20 Variant translocations, seen in the remaining ALK+ ALCL cases, target several partner genes including TPM3/1q21, ATIC/2q35, TFG/3q12, CLTC/17q23, ALO17/17q25, MYH9/22q12, MSN/Xq11 and SEC31A/4q21 (see Table 1). Four fusions, TFG-ALK, MSN-ALK, ALO17-ALK and MYH9- ALK, have been exclusively observed in ALK+ ALCL21 (see Table 1). Given that normal lymphoid cells do not express the ALK protein, immunostaining with ALK-specific antibodies is routinely used for the diagnosis of ALK+ tumours (see Figure 2). Additional diagnostic techniques include conventional cytogenetics, fluorescence in situ hybridisation (FISH) (see Figure 3) and molecular analysis.
Anaplastic Lymphoma Kinase-positive Large B-cell Lymphoma
Subsequent studies showed that ALK expression is not restricted to ALCL but also hallmarks other tumours, including large B-cell lymphoma (LBCL). The first series of ALK-expressing LBCL was reported by Delsol et al.22 in 1997. To date, 54 ALK+ LBCL cases have been described.23–25 The recent WHO classification of haematolymphoid neoplasms considers ALK+ LBCL a distinct lymphoma entity.8 Cytologically, the lymphoma cells display either an immunoblastic or a plasmablastic morphology; occasional binucleated or multinucleated cells mimicking Hodgkin and Reed-Sternberg (H/RS) cells may also be seen.26 ALK+ LBCL shows a distinct immunophenotype, characterised by the strong expression of ALK and EMA and expression of plasma cell markers such as CD138/VS38 and MUM1. Notably, the neoplastic cells have lost expression of B-cell-lineage-associated leukocyte antigens such as CD19, CD20 and CD79a, suggesting terminal plasma cell differentiation.22,23 The disease spans all age groups (nine to 72 years of age, with a median age of 38 years), but approximately 22% of the cases occurred in the paediatric population (<18 years of age). The lymphoma is more common in men than women (M:F ratio of 3:1).23 Most patients with ALK+ LBCL presented with stage III/IV disease and their clinical outcome was worse than ALK+ ALCL, particularly in the paediatric population. Despite aggressive treatment, approximately half of the patients died of disease three to 33 months after therapy. No difference in survival between paediatric and adult cases was found, despite more intensive therapies in the paediatric population.23
Similar to ALK+ ALCL, the aberrant ALK expression in ALK+ LBCL is also driven by 2p23/ALK translocations, of which t(2;17)(p23;q23) is the most frequent (65%). This aberration leads to a chimaeric CLTC-ALK protein and manifests as a distinctive granular cytoplasmic expression of ALK27–35 (see Table 1 and Figure 3). Interestingly, only a few ALK+ LBCL cases (13%) showed the t(2;5)(p23;q35)/NPM1-ALK rearrangement, commonly occurring in ALK+ ALCL25,36–38 (see Table 1). Recently, we have identified a new cryptic SEC31A-ALK fusion in an ALK+ LBCL case generated by complex rearrangements targeting the respective loci at 4q21 and 2p23.25 A functional analysis of the SEC31A-ALK protein and its downstream effectors revealed that this fusion is a constitutively activated tyrosine kinase responding in vitro to the ALK inhibitor NVP-TAE-684.
Anaplastic Lymphoma Kinase Rearrangements in Non-haematological Malignancies
Apart from ALK+ ALCL and LBCL, various tumours of mesenchymal and epithelial origin, including inflammatory myofibroblastic tumour (IMT),39–41 lung cancer42–53 and oesophageal squamous cell carcinoma (SCC),54,55 were found to implicate the ALK gene and inappropriately express the ALK protein (see Table 1). Particularly interesting is the finding of the same translocations and/or ALK fusions in ALK+ malignancies of different origin (see Table 1). TPM3-ALK,56–59 ATIC-ALK,60 CLTC-ALK59,61,62 and SEC31A-ALK63 were reported in lymphomas and IMT, and TPM4-ALK has been found in IMT and SCC.57,59,64 So far, CARS-ALK65,66 and RANBP2-ALK62,67,68 have only been found in IMT, and EML4-ALK and KIF5B-ALK in non-small-cell lung carcinoma (see Table 1).42–53 Recently, oncogenic mutations of the ALK kinase and amplification of ALK have been discovered in neuroblastoma tumours.69–75 In addition, deregulated expression of full-length ALK messenger RNA (mRNA) and/or protein has been described in a variety of primary tumours and cancer cell lines including glioblastoma,76,77 rhabdomyosarcoma,78 Ewing sarcoma and retinoblastoma76 and breast cancer.79 Additional studies are required to assess the possible role of full-length ALK and ALK fusion forms in the pathogenesis and progression of these nonhaematopoietic tumours.
Mechanism of Anaplastic Lymphoma Kinase Fusion Kinase Activation
Under normal conditions, activation of RTKs such as ALK depends on ligand binding. Two putative ALK ligands have been proposed, pleiotrophin (PTN) and midkine (MK),80,81 which show similar expression patterns to ALK (high levels during neural development and decreased expression following birth),82–85 indicating that PTN, MK and ALK could be important for normal neural function. Stoica and co-workers showed that PTN-induced mitogenesis resulted in increased phosphorylation of ALK substrates in ALK-transfected cells,80 and that MK binds to ALK-transfected 32Dcl3 cells.81 Furthermore, ALK is likely to be involved in PTN-stimulated phosphorylation and activation of PI3K, MAPK and PKB/AKT anti-apoptotic proteins.77,86
Molecular studies show that tyrosine-kinase-associated chromosomal translocations in cancer lead to ligand independence of these kinases that then function as constitutively activated tyrosine kinases.With the exception of MSN-ALK and MYH9-ALK, all of the ALK fusion proteins contain the same ALK region, i.e. amino acid residues 1,058–1,620 of the full-length wild-type receptor.87–89 This region contains the complete intracellular part of the receptor, including the tyrosine kinase catalytic domain responsible for the kinase activity (see Figure 1). This C-terminal ALK fragment is fused to the N-terminal end of the partner protein that mediates constitutive self-association of the ALK fusion, mimicking ligand binding. This can be achieved by oligomerisation via a motif such as a coiled-coil domain that is present in the N-terminal portion of the TFG, TPM3, TPM4, MSN, ALO17, RANBP2 and EML4 partner proteins, a nucleoplasmin domain seen in NPM1, WD40-like repeats observed in SEC31A and a di- or multimerisation domain found in ATIC and CLTC, respectively (see Figure 1).90
The subcellular localisation of the ALK fusion protein is dependent on the partner protein. The distribution of NPM1-ALK homodimers is limited to the cytoplasm, but the nuclear localisation motifs present in the wildtype NPM1 permit entry of the NPM1/NPM1-ALK heterodimers to the nucleus and nucleolus that is manifested by the unique nuclear and cytoplasmic ALK immunostaining (see Table 1 and Figure 2).13 The vast majority of variant ALK fusions are localised in the cytoplasm (see Table 1 and Figure 2); notably, CLTC-ALK shows a characteristic granular cytoplasmic distribution (see Figure 2) due to its localisation within the clathrin-coated vesicles of the neoplastic cell. 91 In contrast, the MSN-ALK fusion is expressed at the cell membrane, probably because of the association of moesin with other cell membrane proteins. 92
Mouse Models of Nucleophosmin–Anaplastic Lymphoma Kinase-induced Lymphoma
In this article, we focus on the mouse models of NPM1-ALK-induced lymphoma that were the main focus of investigations. Of note, however, a few variant fusions have also been used to generate transgenic mice, i.e. TPM-ALK93 and EML4-ALK.49,94
Chimaeric Mouse Models
The initial study investigating the tumorigenic capacity of NPM1-ALK in vivo was performed by Kuefer et al.95 The established NPM1-ALK bone marrow transplant mouse model resulted in the development of B-lymphoid immunoblastic tumours with a relatively long latency (four to six months) and moderate penetrance of tumour development. This phenotype was unexpected, as at that time the only known human ALK+ lymphomas were T-/null-cell ALCL. In subsequent retroviral transduction experiments, Miething et al.96 used retroviral constructs with a promoter that drives higher expression of the chimeric kinase. When bone marrow was transfected with high retroviral titres, the mice developed fatal haematopoietic malignancies possessing a myeloid/ macrophage cell phenotype with a short latency (three to four weeks). The use of low titres of retrovirus gave rise to the development of B-lymphoid tumours with a longer latency (12–16 weeks). Overall, there seems to be a relationship between the degree of induced NPM1-ALK expression and the phenotype of the resulting tumour, with higher levels of NPM1-ALK expression leading to a myeloid/macrophage malignancy and lower levels in B-cell lymphoid tumours.
To generate experimental T-lymphoid malignancies, Miething and co-workers used Cre-conditional NPM1-ALK retrovirus to transduce bone marrow from two lines of Cre-transgenic mice.97 Lysozyme M-promoter Cre mice expressed Cre recombinase from the myeloid-specific lysozyme M promoter and developed a fatal histiocytic malignancy after four to six weeks. Granzyme B-promoter mice expressed Cre recombinase from the T-cell active granzyme B gene promoter and showed a mixed T-NHL/histiocytic phenotype with the same latency (four to six weeks).
Transgenic Mouse Models
Several groups have reported transgenic NPM1-ALK mouse models in which expression of the fusion is driven from different promoters. In one transgenic CD4-NPM1-ALK mouse line in which NPM1-ALK expression is driven by the T-cell-specific CD4 promoter, CD30+ T-cell lymphomas arose, whereas a second line showed tumours with a plasma cell phenotype, suggesting leakiness of the CD4 promoter.98 Use of the haematopoietic-cell-specific Vav-promoter and the T-cell-specific CD2-promoter to express NPM1-ALK in transgenic mice resulted in both the development of B-cell tumours, more specifically plasmacytomas (high-copy-number mice) and putative peritoneal B1- cell-derived tumours (low-copy-number mice) in Vav-NPM1-ALK mice,99 and diffuse large B-cell lymphoma and histiocyte/T-cell-rich B-cell lymphoma in CD2-NPM1-ALK mice.100 All three models showed a long latency ranging from four to five months (CD4-NPM1-ALK) to three to nine months (Vav-NPM1-ALK) and more (CD2-NPM1-ALK). In contrast, Lck-NPM1-ALK mice, with NPM1-ALK expression from theT-cell specific Lck gene promoter, developed large T-cell lymphoblastic lymphomas after only eight weeks.101
Xenograft Mouse Models
Several studies have been published in which xenograft mouse models of ALK-mediated tumorigenesis are described. JB6, COST, Karpas-299 and Ba/F3 cells expressing NPM1-ALK were used to test whether tumours formed in immunocompromised severe combined immunodeficiency (SCID) mice. Localised subcutaneous xenografted tumours and systemic lymphomatous disease could be established in all SCID mouse models.102–105 In addition, the mice benefited from treatment with ALK inhibitors, suggesting ALK is a good therapeutic target.
Anaplastic Lymphoma Kinase Signalling in Cancer
Research on ALK signalling in cancer has mainly been focused on the effect of NPM1-ALK on signalling pathways. The observation that NPM1 and almost all variant partners of ALK contain some form of an oligomerisation motif suggests that the mechanism underlying ALK tyrosine activation and the activated signal transduction pathways are probably similar for all ALK chimaeric proteins.106
The oncogenic potential of ALK fusions translates into enhanced cell proliferation and survival, cytoskeletal rearrangements and changes in cell shape.95,98,107,108 The aberrant tyrosine kinase activity triggers several different pathways that are interconnected and overlapping. The rat sarcoma viral oncogene homologue (RAS)/mitogen-activated protein kinase (MAPK) and phospholipase C-γ (PLC-γ) pathways are important in mitogenesis, whereas phosphatidylinositol 3-kinase (PI3K/AKT) and STAT pathways are of primordial importance in cell survival and phenotypic changes. Tandem mass spectrometry has identified 46 putative NPM1- ALK-interacting proteins, suggesting the NPM1-ALK signalosome may be considerably more complex than previously assumed.109
Enhancement of Proliferation
Activation of the RAS/extracellular signal-regulated kinase (ERK) pathway is the main cause of the increased growth of ALCL cells. In these cells, NPM1-ALK acts as a docking molecule for several downstream adaptors or scaffolding molecules, such as insulin receptor substrate 1 (IRS1), SRC homology 2 domain-containing transforming protein (SHC) and growth factor receptor-bound protein 2 (GRB2), which bind to specific autophosphorylated tyrosine residues of ALK and induce the RAS/ERK pathway.92,110 Recently, it has been shown that NPM1-ALKactivated tyrosine phosphatase SHP2 stimulates ALCL growth through ERK1/2 phosphorylation.111 Also, NPM1-ALK-mediated mammalian target of rapamycin (mTOR) activation has been demonstrated to be transduced by the RAS/ERK signal transduction pathway.112 Activation of the PLC-γ pathway is also thought to play an important role in mitogenesis induced by ALK signalling. PLC-γ binds directly to NPM1-ALK via interaction between the SH2 domain of PLC-γ and tyrosine residue Tyr664 of NPM1-ALK. Mouse Ba/F3 and Rat 1a cells expressing a mutant NPM1-ALK that cannot phosphorylate Tyr664 were unable to bind and activate PLC-γ, emphasising the importance of NPM1-ALK Tyr664.113
Studies indicate that NPM1-ALK interacts with a nuclear-interacting partner of ALK (NIPA), initially described as an anti-apoptotic protein.114 More recent studies, however, consider NIPA as an F-box-containing protein controlling mitotic entry115 that is involved in cyclin B1 proteasome degradation.116 Approximately 50% of ALCL cases show loss of the tyrosine phosphatase SHP1 that acts as a tumour suppressor gene via inactivation of the JAK3-STAT3 pathway, the abrogation of which usually correlates with uncontrolled cell growth.117,118
Levantaki et al. showed that expression of NPM1-ALK induced phosphorylation of JUN N-terminal kinase (JNK) and cJun was linked to a dramatic increase of activator protein 1 (AP-1)-transcription-factor activity. This may contribute to uncontrolled cell cycle progression and oncogenesis.119
Regulation of Apoptosis
Apart from an increase in cell proliferation, cancer cells need to suppress apoptosis in order to become tumorigenic. NPM1-ALK has been shown to activate PI3K in ALCL through binding to its regulatory p85 subunit, resulting in phosphorylation of the serine/threonine kinase AKT.120,121 AKT then phosphorylates the pro-apoptotic protein BAD, leading to its dissociation from the anti-apoptotic protein BC-XL, which can then block cell death.122,123 The anti-apoptotic (and proliferation-enhancing) effect of NPM1-ALK via AKT can also, in part, be mediated through AKT-induced inhibition of forkhead family transcription factor (FOXO3a) activity, promoting cell-cycle progression.124,125 In addition, PI3K/AKT activation induces phosphorylation of the cyclin-dependent kinase inhibitor 27, increasing its proteasomal degradation and promoting cell-cycle progression.126 Recent evidence points to a role of the PI3K/AKT pathway to cause constitutive activation of the dual specificity phosphatase CDC25A, leading to enhanced proliferation.127 Also, a positive synergistic effect between the sonic hedgehog (SHH)/GLI1 and PI3K/AKT pathways has been described that contributes to the lymphomagenic effect of NPM1-ALK.128
The activated phosphorylated form of the STAT3 transcription factor that has been detected in 84% of ALK+ ALCL tumour samples, as well as in cell lines and in thymocytes and tumour tissues of NPM1-ALK transgenic mice98,129–133 leads to the enhanced expression of anti-apoptotic proteins, cell-cycle regulators and immunosuppressive proteins such as BCL-XL, survivin, cyclin D3, C/EBPb, myeloid leukaemia sequence 1 (MCL1) and PD-L1 (CD274).132,134–136 A pro-proliferative and anti-apoptotic role for STAT5B in NPM1-ALK-mediated signalling has been described by Nieborowska-Skorska and colleagues.137 However, later studies showed that this mechanism of activation requires JAK21.38 In contrast, Zhang et al.139 suggested that STAT5 acts as a tumour suppressor in ALCL as it is epigenetically silenced by the NPM1- ALK tyrosine kinase, but when re-expressed it reciprocally inhibits NPM1-ALK signalling.
Cell Migration and Cytoskeletal Rearrangements
In vitro studies showed that various ALK fusions increase the migration rate of lymphoid cells and change their shape,107,111 and that lymphoid cells become anaplastic in the presence of transforming ALK proteins.140 Notably, inhibition of ALK-kinase activity by small-molecule inhibitors dramatically reduces ALCL-cell motility and reverts morphology from anaplastic to spherical.141 Several lines of evidence indicate that NPM1- ALK activates proteins involved in the regulation of actin filament assembly and cytoskeletal rearrangements, such as p130Cas,107 SHP2,111 pp60Src,142 Rac1143 and Cdc42,144 influencing the morphology, migration and transformation of ALK-expressing tumour cells.
Modulation of the Anaplastic Large Cell Lymphoma Immunophenotype
ALCL is characterised by high expression of CD30, which is also strongly expressed by the neoplastic (H/RS) cells of Hodgkin’s lymphoma (HL).18 Transduction of NPM1-ALK into HL-derived cell lines induces an ALCL-like phenotype and morphology. These observations suggest a role for CD30 in the development of the ALCL phenotype.140 In addition, NPM1- ALK impedes CD30 signalling and nuclear factor kappa-B (NF-κB) activation dependent on both ALK kinase activity and the N-terminal NPM1 domain.140
Treatment
Both ALK-expressing ALCL and LBCL are conventionally treated by combination chemotherapy. In a minor group of ALK+ LBCL patients, chemotherapy in combination with radiotherapy (15%) or radiotherapy alone (2%) was given.23 ALK is a strong candidate for targeted treatments. As numerous malignancies showing aberrant ALK expression have been reported, the search for new ALK inhibitors is rising.145–147 Recently, several small-molecule inhibitors displaying selective and potent ALK inhibition in both in vitro tumour models and in vivo xenograft ALCL-tumour models have been described, among them, NVP-TAE-684, fused pyrrolocarbazole (FP)-derived small molecules (CEP-14083 and CEP-14513) and PF-2341066.25,102,148,149 Inhibition of NPM1- ALK as well as SEC31A-ALK-expressing Ba/F3 cells by NVP-TAE-684 suggests that it can be used in the therapy of patients with both classic and variant ALK fusions25 (see Figure 4). Given that virtually all ALCLs express CD30, this molecule is a promising target for therapy with anti- CD30 monoclonal antibodies. A recent phase I multidose study of SGN- 30, a chimaeric monoclonal antibody for the treatment of CD30(+) malignancies, holds promising results.150
Recent studies demonstrated that ALK is spontaneously immunogenic in ALCL cells resulting in B-cell, cytotoxic T-cell (CTL) and CD4 T-helper response.151–154 Chiarle and colleagues155 showed that DNA vaccination with plasmids encoding portions of the cytoplasmic ALK domain protects mice from local and systemic lymphoma growth; hence, ALK vaccination following chemotherapy and/or pharmacological inhibition of ALK can be an interesting approach to treat ALK-expressing malignancies, such as ALK+ ALCL and ALK+ LBCL. Genetic targeting of ALK through small interfering RNA (si)RNA and single-stranded antisense oligodeoxynucleotides (ASO) may become important in the future; however, in vivo delivery of these molecules remains an unsolved problem.156 Finally, molecules acting downstream of ALK signalling, such as PI3K/AKT, JAK/STAT3, PLC-γ and RAS/MAPK, can also be potential targets of selective inhibition. However, because several signalling cascades are interconnected and are not dependent only on ALK signalling, it remains to be elucidated whether targeting these proteins will result in tumour eradication. ■