Breast cancer is the most common malignant disease in women: according to US statistics, one in eight women will be diagnosed with breast cancer in her lifetime.1 It was estimated that approximately 232,340 new cases of invasive breast cancer and 39,620 deaths were expected among US women in 2013.2 Early detection and diagnosis of breast cancer are essential for successful treatment; women diagnosed with stage II and III breast cancer have a high risk for recurrence and a higher chance of developing metastatic disease, which remains incurable.3 Early diagnosis of breast cancer is associated with significantly lower morbidity; tumor detection at stages 0 and 1 is associated with approximately 98 % 5-year survival.4
The study of biomarkers in the monitoring of tumor progression began with the discovery of carcinoembryonic antigen (CEA) in 1965.5 Biomarkers are potentially useful in screening programs, but the application of biomarker testing requires the incorporation of radiologic screening methods in order to locate the tumor for further treatment/assessment. Biomarkers can also be used in the diagnostic workup of suspicious lesions as part of the clinical decision-making process, and may provide biochemical evidence to inform decisions regarding the need for biopsy, much like positron emission tomography and computerized tomography (PET-CT) that measures the metabolic rate of a mass in conjunction with its anatomical presentation. The third potential use is to monitor for breast cancer disease recurrence. This article will outline the limitations of current breast cancer diagnostic methods and examine the role of biomarkers in current screening paradigms.
Limitations of Current Breast Cancer Screening Techniques and the Potential Role of Biomarkers
Current screening in the US for breast cancer relies heavily on mammography, which is an oversold and expensive regimen that detects only 70 % of breast cancers,6 while studies of multimodality imaging suggest that mammographic sensitivity is below 50 %.7 Mammographic screening is less effective in younger women. In 2009, the US Preventive Services Task Force (USPSTF) gave mammography a ‘C’ recommendation (routine screening not recommended) for women aged 40–49,8 largely because most diagnostic pathways are associated with false positives, causing anxiety and increased expense. The USPSTF suggested a ‘C’ recommendation despite the fact that their meta-analysis demonstrated a 15 % relative reduction in mortality with screening in this age group; they considered the harmful effects to be greater than the relatively small absolute benefit. The most recent update of the Canadian National Breast Screening Study presented a pessimistic scenario of mammographic screening. Annual mammography in women aged 40–59 did not reduce mortality from breast cancer beyond that of physical examination or usual care when adjuvant therapy for breast cancer was freely available.9 Importantly, Canadian data prior to this recent study were included in the 2009 meta-analysis performed by the USPSTF. This meta-analysis concluded that most of the historic screening trials showed a benefit to screening women aged 40–49, overriding the ‘no benefit’ statistical influence of the Canadian trial. It should be noted that the Canadian trial has been controversial in the community due to corruption of the randomization process and inclusion of palpable masses in the mammography trial.
Imaging is also confounded by individual patient characteristics, such as breast density, especially in women under the age of 50. Indeed, more than 19 states have passed or are considering legislation that would require disclaimers as to imaging effectiveness in women with dense breast. This group of women could be particularly benefitted by an additional nonimaging-based approach, such as the use of biomarkers.
A larger body of evidence supports the use of mammographic screening in women aged 50 and over. A recent prospective cohort study of a national screening program in all Norwegian women aged 50–79 between 1986 and 2009 concluded that invitation to modern mammography screening reduces breast cancer mortality by 28 %,10 which is similar to the 20 % mortality reduction described in a recent meta-analysis of 11 historic randomized trials.11 The study concluded that screening reduces breast cancer mortality but that some overdiagnosis occurs, although estimates of its magnitude are unreliable.
Widespread mammography has resulted in an increase in the detection of early-stage disease, (stage 0 and 1 cancers). However, incidence of stage II and III disease has not fallen proportionately, suggesting a bias in the detection of indolent cancers rather than aggressive cancers.12 It is important to distinguish between early diagnosis in general and mammographic screening diagnoses. While the importance of mammography seems evident, early diagnosis that includes cancers currently missed by mammography could accomplish more. Breast cancer biology appears to be much more vulnerable to early detection than is currently being demonstrated with clinical trials using mammography alone. The reasons for this assertion will be discussed below.
First, prospective, randomized trials understate the power of early diagnosis because the data are derived from ‘invitation to screen’ rather than from those patients who actually complied with the protocol. In other words, the ‘intention-to-treat principle,’ which is considered a ground rule for prospective randomized trials, seriously underestimates the benefit of early diagnosis with mammography when patient compliance is low. In addition, during lengthy clinical trials of screening mammography, patient compliance can be marginal, with women in the ‘no mammography’ groups having mammograms performed anyway, and vice versa. When outcomes are published based on compliant patients only, it is interesting that the mortality reduction is greater than in the official trial results, an indirect indicator that the benefit of mammography is being understated. Second, mammographic sensitivity is routinely overstated. With a more realistic sensitivity in the range of 50 %, taken from the multimodality studies, the current mortality reduction demonstrated in most clinical trials reflects the diagnosis of only half of the detectable cancers (if other modalities had been used). Good sensitivity of the screening tool coupled with underlying cancer biology that is vulnerable to early detection forms the core of successful screening. Thus, if sensitivity of the screening tool is low, the biologic vulnerability to early detection must be high if a mortality reduction has been demonstrated. Therefore, the proposed basis for improving early diagnosis, is the detection of breast cancers currently missed by mammography.
Breast magnetic resonance imaging (MRI) is much more sensitive than mammography. In a study of 1,387 women of average risk, pre-screened with examination, mammography, and 89 % with ultrasound, a single MRI revealed that 1.1 % of the women had breast cancer, a yield greater than most mammography screening programs.13 A study of a 1.5-T breast MRI system found a sensitivity of 92 % and specificity of 89 %.14 However, MRI is too expensive for routine screening. In order to be cost-effective, it has been calculated that a breast cancer prevalence of 3 % is needed.15 Cancer yields for MRI screening trials to date have been 2.2–3.6%.16–21
Risk stratification has been suggested in selecting which women should undergo MRI screening. Current guidelines by the American Cancer Society (ACS) and National Comprehensive Cancer Network (NCCN) base screening recommendations on lifetime risks.22,23 However, risk stratification is only one of two predictors of occult cancer; the other is breast density. Prior biopsies and surgery, breast augmentation, and other factors can also obscure breast images in the general population. In highrisk patients, multimodality imaging has revealed low sensitivity (40 % sensitivity overall) when mammograms are used alone.7By using breast density as an equal predictor to traditional risk, a study has revealed a 2 % probability of finding occult cancer with MRI in a patient with a normal mammogram.24 In the American College of Radiology Imaging Network (ACRIN) 6666 study of 2,809 women with elevated cancer risk and dense breasts, the addition of whole breast screening ultrasound to mammograms increased the overall sensitivity of cancer detection, yet the combined sensitivity of this dual modality approach was still suboptimal after three screening sessions when a single-breast MRI was performed at the completion of the study.25
A better approach would be to select patients for auxiliary imaging based on biomarkers rather than through risk stratification. Mathematical modeling, using a biomarker test with reasonable accuracy, predicts cancer yields with MRI well above that being currently achieved by relying on risk levels. Biomarkers should theoretically indicate the presence of occult cancer in patients with a normal mammogram, a more-specific method than calculating lifetime risk. A biomarker-based screening paradigm would involve a blood test as the initial screen, with auxiliary imaging used to confirm and localize the cancer.
Integration of Biomarkers into Current Diagnostic Paradigms
The American College of Radiology Breast Imaging Reporting and Data System (BIRADS®) has been established as a standardized means of reporting mammography results, with implications for diagnostic workup.26 The original version of BIRADS had a very wide range of cancer probability (3 % to 89 %) for Level 4 interpretations, wherein biopsy was recommended for all. While an overall probability could be calculated,27 an individual positive predictive value (PPV) was not specific until the adoption of the BIRADS 4a, 4b, 4c system, which provided PPVs that were more useful clinically. However, the BIRADS system has limitations, with radiologists differing in their assessments of BIRADS 3 (no biopsy; 6-month follow-up) versus BIRADS 4a (biopsy recommended). Radiologists frequently have to make subjective decisions about whether to perform biopsies, and the majority of these breast biopsies are still negative, even after several decades of experience with mammography. A biomarker with a strong negative predictive value (NPV) could potentially avoid the need for biopsy in BIRADs 3/4a lesions. Likewise, a biomarker with a strong PPV may indicate the need for biopsy in BIRADS 3/4a lesions (some cancers e.g. medullary, papillary, and mucinous, can appear benign on imaging). Biomarkers therefore have the potential to improve the cost-effectiveness of diagnosis.
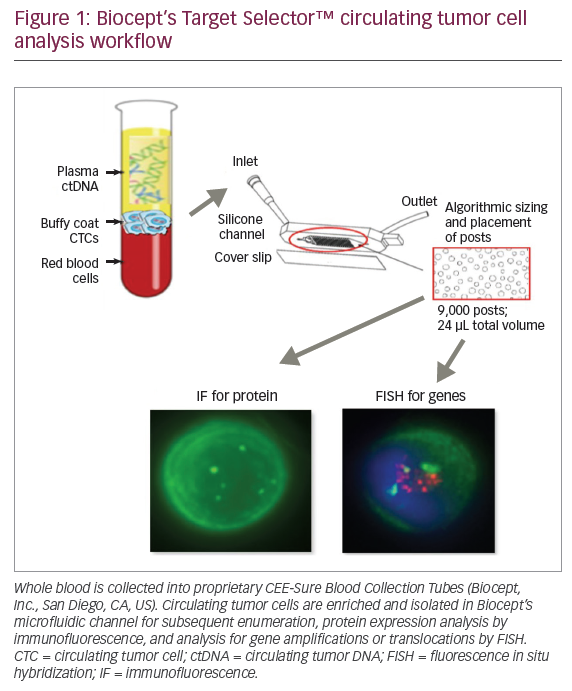
In high-risk patients, multimodality imaging is already employed, or at least recommended, and biomarkers are unlikely to add useful data. If a biomarker assay is performed at the same time as the MRI, a normal MRI but abnormal biomarker result could be confusing. One scenario to explain abnormal biomarkers with a negative MRI would be subclinical disease, not yet detectable by any known imaging method. Short interval follow-up with MRI (and other imaging methods) could be proposed, but if cancer yields in this scenario are negligible, the biomarkers would need to be considered false positive. That said, high cancer yields with short-term follow-up would be true positives, and the benefit of ‘earlier’ detection would need to be assessed. A suggested screening protocol incorporating the use of biomarkers is given in Figure 1.
Biomarkers for Disease Monitoring and Therapeutic Decision-making
Biomarkers should theoretically be a useful indicator of systemic recurrence; however, to date, the earlier detection of systemic disease achieved with biomarkers has not improved outcomes.28 Existing biomarkers have largely proved disappointing because they lack sensitivity and specificity, and also tend to detect metastatic disease rather than early-stage breast cancer.29 While it may be intuitive to believe that any biomarker test that detects localized disease in the breast will, by its very nature, be helpful in the detection of metastatic disease, this may not be the case. Specific biomarkers might herald early carcinogenesis, but then be suppressed (nondetectable) later in the natural evolution of progressive disease. Currently, the major clinical application of serum biomarkers is in monitoring therapy in patients with advanced breast cancer that is not assessable by existing clinical or radiologic procedures.28 One such example is CA27.29, which is approved by the US Food and Drug Administration (FDA) for the monitoring of breast cancer patients after diagnosis and treatment.
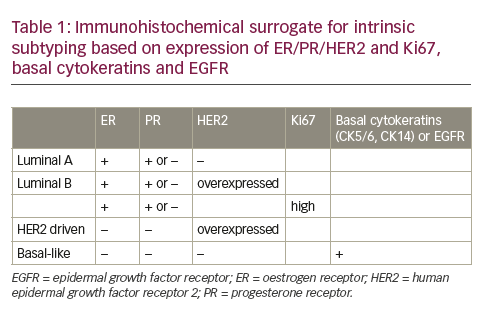
Much of the success in the use of diagnostic tests for breast cancer management and monitoring has come from assays designed to assist in patient prognosis and treatment decisions. Tests such as OncotypeDx,30 Prosigna,31 and MammaPrint32,33 are all tests that provide either prognostic information or treatment decision support (see Table 1). These tests require breast tumor sample and therefore are primarily designed to provide predictive information based on patient-specific information. While not useful in the detection of cancer, these tests have nonetheless changed the way clinicians use molecular tools in the practice of medicine.
Potential Biomarkers in Breast Cancer Screening
The pathogenesis of breast cancer involves the disruption of the breast structure. Proliferation of epithelial cells, loss of acinar structure, and filling of luminal space are commonly seen during premalignant stages of breast cancer and in lesions such as ductal carcinoma in situ (DCIS). The transformation to invasive disease is associated with the loss of tumor suppressor function and/or the gain of oncoproteins, which regulate many downstream pathways. Identification and characterization of candidate DNAs (cDNAs) and proteins involved in these oncogenic transitions is central to the development of biomarker assays. Some studies have correlated breast cancer prognosis with proteins in the tumor, such as hormone receptors, human epidermal growth factor receptor (HER2), and proliferating cell nuclear antigen (PCNA).28,34,35 However, accessing these proteins requires biopsies, which is ‘after the fact’ when discussing screening or diagnostic work-ups. Studies have therefore focused on developing serum-based assays to quantify soluble molecules released into the bloodstream by cancer cells or other cell types belonging to the tumor microenvironment. A summary of discovery strategies for biomarkers in breast cancer is provided in Figure 2.
Many proteins have been identified as potential serum biomarkers in breast cancer. CEA is the most studied of these,36 but in studies of its single use for diagnosis of early breast cancer, it was too insensitive and nonspecific to reliably differentiate patients with early breast cancer from those with benign disease or disease-free.37 Indeed, CEA identifies cancers other than breast, lending to its lack of specificity as a single biomarker. Cancer antigen 15-3 (CA15-3) is the soluble form of mucin 1 (MUC-1). Early studies suggested that it had limited sensitivity in the diagnosis of breast cancer,28 but a recent study evaluating the value of CA15-3 for the diagnostic integration of molecular imaging findings performed with hybrid PET/CT technology, found that serial increases in CA15-3 could be used to predict positive PET/CT results.38
Following early clinical data supporting the use of the tumor marker CA27.29 in monitoring therapeutic response,39 the Simultaneous Study of Docetaxel-Gemcitabine Combination Adjuvant Treatment, as well as Extended Biphosphate and Surveillance (SUCCESS) clinical trial evaluated the role of CA27.29 before and after adjuvant chemotherapy, as well as after 2 and 5 years in patients with early breast cancer. No correlations were found between CA27.29 and nodal status, grading, hormonal status, and HER2/neu status on the primary tumor. However, larger tumor size, lobular histology, older age, and postmenopausal hormone status before the start of treatment were significantly associated with higher CA27.29 levels.40 However, it is of limited utility as a biomarker in breast cancer screening; its utility is as a prognostic indicator of early recurrence. In addition, in the SUCCESS trial, circulating tumor cell (CTC) counts were positively correlated with outcome and prognosis, however, to date, these results have not altered clinical practice to a large degree. Soluble HER2 may have limited utility in breast cancer screening;41 it is also useful for monitoring treatment response.42 In a study of 30 newly diagnosed patients with breast cancer, an oncogenic protein that regulates RNA-protein interaction (RS/DJ-1) was readily detectable in sera from 37 % of newly diagnosed patients but not in controls (116 patients with other cancers and from 25 healthy subjects).43 Other proteins have demonstrated utility as prognostic indicators. These include estrogen receptor (ER) and progesterone receptor (PR)44 urokinase-type plasminogen activator (uPA) and its inhibitor plasminogen activator inhibitor-1 (PAI-1).45
Biomarkers of the immune system also offer potential in breast cancerscreening. Two instances implicate the immune system as a prime target for early breast cancer biomarkers. First, during breast carcinogenesis, antigen upregulation, mutation, altered glycosylation, and altered degradation are seen and these changes lead to an amplification of the immune response. Additionally, host response to the tumor can result in the production of autoantibodies. These antibodies are often produced to wild-type cancer proteins that are functionally misregulated. Local immune responses to tumor antigens are amplified in draining lymph nodes, and then enter the systemic circulation.46 Autoantibodies generated against tumor antigens can be detected in the sera of cancer patients and are potentially useful screening biomarkers as they are stable, specific, may be present in higher concentrations than their cognate antigens, easily purified, and readily detected with validated tests. They have also been found to correlate with tumor burden and disease progression.47 However, they have poor sensitivity as single analytes, and differences in protein purification and assay characteristics have limited their clinical application.48,49 Two serum autoantibodies, HER2/neu and p53, have been detected in sera collected more than 150 days before a breast cancer diagnosis.50 However, p53 autoantibodies can only be detected in the sera of up to 20 % of patients with breast cancer. Levels of p53 autoantibodies depend on numerous factors, including tumor burden, expression level, and p53 mutation. Other autoantibodies with limited utility as single agents include NY-ESO-1,51 SCP-148, and SSX2.49
Recently, panels of autoantibodies discovered by Drs Anderson and LaBaer have led to the exciting prospect of a screening test that incorporates many autoantibodies, each with a specificity of 80–90 %.39 In order to increase the predictive value of tumor-specific autoantibodies for use as immunodiagnostics, this research has focused on testing multiple antigens in parallel.52,53 Antibodies to hundreds of other tumor antigens have been identified in the sera of breast cancer patients, and may correlate with disease progression.54,55 However, testing of these panels in prospective populations of patients and validating this approach is critical.
There is an increasing focus on the use of CTCs rather than biomarkers, prompting the hypothesis that CTCs may be used as biomarkers.56 However, they are currently being investigated only as markers of prognosis, and their utility in screening assays is unknown. The primary difficulty in using CTCs is that their enrichment in detection protocols has largely depended on single or duplex markers. Tumor heterogeneity makes this approach less attractive because of the lack of any two unifying proteins to detect tumors. In addition, CTCs are present at such a low quantity that the challenges for their use in a clinical setting are anything but trivial.
Cytokine-based assays have shown discrepant results. One study found no significant difference in the baseline serum interleukin-8 (IL-8) and IL-12 p40/p70 levels between breast cancer patients and healthy controls57 while another noted levels of IL-8 increased in advanced disease and increased with disease progression in women with breast cancer.58 Indeed, IL-8 has been shown to be a critical mediator of epithelial to mesenchymal transition, a required step for tumor spread. A more recent study of a cytokine based assay IL-8, IL-12, vascular endothelial growth factor (VEGF), and hepatocyte growth factor (HGF), revealed age-dependent shifts in cytokine expression measurement,59 an important consideration for future development of cytokine-based assay. This observation is likely due to the fact that in women, menopause drastically changes their hormonal make-up. Since many of these factors are regulated by hormone levels, developing a baseline for women of all ages is problematic.
Biomarker Microarray Techniques In Breast Cancer Detection
The methods of determination of known proteins in multiple arrays simultaneously, form the basis of microarray techniques. Protein microarrays have considerable potential as diagnostic tests as they have the capacity for rapid screening of numerous analytes and require only a few microliters of serum per assay.60 These assays match the needs in the clinic as clinicians require real-time measurements to determine the best course of action for a given patient. Known tumor antigens as well as predicted tumor antigens can be included to create a comprehensive protein tumor antigen array. However, they have been associated with difficulties in production and validation, and early proteomics-based approaches to distinguish between the sera of those with cancer from healthy sera were hampered by the difficulty of identifying small quantities of protein fragments within complex mixtures, as well as protein instability and natural variations in protein content of sera within patient populations.61,62
Protein microarrays are available in two basic formats. In forward phase arrays, capture reagents, including autoantibodies and proteins, are arrayed at defined positions on a immobilizing substrate, which is then interrogated with a variety of probes, from protein lysate and enzymes to small molecules and nucleic acids. In reverse phase arrays, protein lysates are spotted. The arrays are then probed with antibodies to the proteins, one antibody probing multiple samples as opposed to one sample for multiple capture reagents.
Programmable protein microarray enzyme-linked immunosorbent assays (ELISAs) have been adapted for the detection of autoantibodies in breast cancer sera.39 A novel protein microarray technology, the Nucleic Acid Protein Programmable Array (NAPPA), spots protein-coding cDNAs rather than proteins and has been developed for the detection of tumor specific autoantibodies in sera from patients with early-stage breast cancer (IBC), and bound immunoglobulin G (IgG).47 The spots, containing specific cDNAs, are used as templates from which protein can be made directly on the array. From 4,988 candidate tumor antigens, 28 antigens were identified as potential biomarkers for the early detection of breast cancer and confirmed using an independent serum cohort. A classifier was identified with a sensitivity of 80.8 % and a specificity of 61.6 % for the entire group of 28 antigens.
The dtectDx™ Breast Serum Biomarker Test
The dtectDx™ Breast (Provista Diagnostics) serum biomarker test analyzes concentrations of five serum biomarkers: IL-8, IL-12, VEGF, CEA, and HGF via ELISA (see Figure 3). These data are combined with select patient characteristics in an algorithm that generates a value of normal or elevated risk.
A clinical study evaluated the clinical performance of the dtectDx Breast test in 898 women aged 35–49 and compared the results with previously published data.59 The assay performed well, correctly identifying four of four invasive cancers and four of five cases of DCIS. The sensitivity was 88.9 % and specificity was 86.7 %, although the false positive rate was higher than expected (13.9 % versus 6.8 %).63 Furthermore, two of the invasive cancer cases were not detected using standard screening methods. The assay may be particularly useful in younger women and those with dense breasts: one study participant was a 38-year-old woman with heterogeneously dense breasts, but otherwise normal on mammography and a normal ultrasound. However, the dtectDx assay identified this patient as being at elevated risk. An MRI led to the discovery of a 2 mm lesion, and a biopsy revealed well-differentiated invasive ductal carcinoma. Additionally, another participant was identified as being elevated by the dtectDx assay while a Dx mammogram provided a BIRADS 2 benign assessment. An MRI was requested due to the 42-year-old women’s breast density and revealed a 1.5 mm lesion. A biopsy later confirmed invasive ductal carcinoma + DCIS.
While these results are compelling for women under the age of 50, the majority of women will be diagnosed with breast cancer after this age. In this cohort, the dtectDx assay performance is not as robust. For this reason, a panel of 10 protein biomarkers in combination with autoantibodies has been developed to detect breast cancer in women of all ages. To validate this next-generation platform, a prospective clinical trial has enrolled 351 women with a suspicious lesion and aims to establish an acceptable algorithm for generation of a single numerical score for women under the age of 50.64 This assay, composed of serum autoantibodies that generate high specificity and low sensitivity, will be paired with protein markers that are low specificity and high sensitivity. A second prospective clinical study Provista-002 is currently enrolling 500 patients with the intent to establish a numerical score cut-off that differentiates malignant from nonmalignant breast cancer in women age 25–75, as well as assessing the overall efficacy of the next-generation test.65
In summary, in women under 50 in a commercial setting, the dtectDx Breast assay improves the detection of breast cancer. Ongoing development has resulted in a test that is highly promising in the ability to detect breast cancer in a broader population of women in conjunction with standard imaging practices. Further clinical development is required before serum biomarkers may be incorporated into routine clinical practice, but the Dx Breast assay represents a significant clinical advance.
Summary and Concluding Remarks
Blood-based biomarker tests offer the potential to improve the sensitivity of breast cancer screening over mammography alone, prompting clinicians to employ multimodal imaging. Potential biomarkers include serum protein biomarkers (SPBs) and tumor-associated autoantibodies (TAbs). The latter offer the greatest potential in screening assays as they are highly specific, biochemically stable in blood and in general, and correlate with tumor burden and disease progression. Many routinely used serum biomarkers have limited use in screening assays as single agents due to their low sensitivity and inability to distinguish between benign and malignant lesions. However, the use of protein microarrays that detect multiple autoantibodies and proteins offer enhanced sensitivity. The dtectDx Breast serum biomarker test comprises a combination of five serum biomarkers for breast cancer. Clinical validation of this assay showed high sensitivity and specificity. The next generation of this test has since been developed, and features protein biomarkers, including autoantibodies, which should improve the clinical utility of the assay. The results of both the 351 and 500 patient prospective randomized studies are designed to demonstrate the validity of the new test. It is important to emphasize that serum biomarkers are currently not the standard of care and are not recommended in any guidelines. In order to become incorporated into routine clinical practice, large clinical trials are required. However, the use of biochemical evidence to improve performance in the detection of breast cancer by imaging will clearly represent a significant step toward precision medicine.