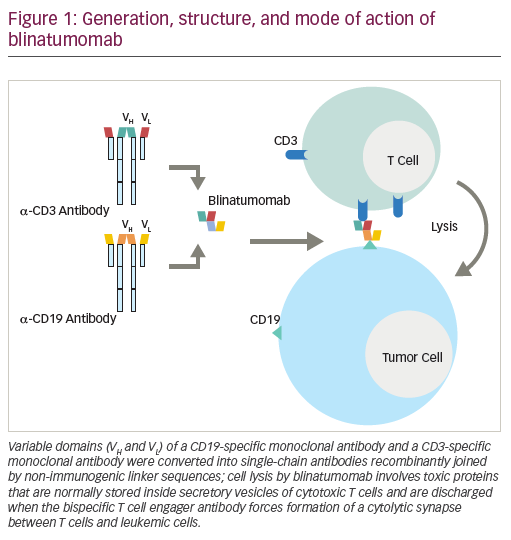
Progress in culturing techniques, including the use of B-cell mitogens in combination with conventional cytogenetics, led to the discovery of non-random chromosomal abnormalities in chronic lymphocytic leukaemia (CLL), initially reported in 1980.1–3 Due to the low in vitro mitotic activity of CLL cells, chromosomal banding had previously proved difficult. One decade later, in 1990, a comprehensive study of clonal chromosomal aberrations revealed an association between specific chromosomal abnormalities and clinical outcome in CLL.4 Here, genomic aberrations were detected in approximately 50% of the patients studied, and for the first time prognostic subgroups in CLL were defined based on chromosomal abnormalities. Trisomy 12 was found to be the most frequent aberration and was associated with poor survival, while patients with a deletion in 13q showed comparable survival to those with a normal karyotype. Taking advantage of improvements in molecular cytogenetics, Döhner et al. used fluorescence in situ hybridisation (FISH) analysis and were able to detect chromosomal abnormalities in more than 80% of CLL patients in a breakthrough publication in 2000.5 The most frequent changes detected were deletions in 13q, 11q and 17p, as well as trisomy 12, with the majority of patients showing single aberrations only. Importantly, deletions in 11q and in particular 17p were associated with poor clinical outcome, while, by contrast, patients with deletion of 13q as a sole abnormality were found to have a favourable clinical course. Indeed, genomic aberrations were shown in many follow-up studies to be important independent predictors of clinical outcome in multivariate analysis, regardless of clinical stage.6,7 Today, FISH analysis of interphase nuclei is the gold standard for detection of recurrent genomic aberrations and is applied in routine clinical diagnostics. However, new technologies including various microarray-based technologies are becoming increasingly popular in screening for novel aberrations. In this review, the latest findings in recurrent genetic alterations, including translocations, and their pathological and prognostic relevance in CLL will be discussed.
Recurrent Alterations in Chronic Lymphocytic Leukaemia
Deletion of 13q14
As mentioned above, structural aberrations involving 13q in CLL were initially reported more than 20 years ago.8,9 Today, deletion of 13q is the most commonly found structural genetic aberration in CLL, detected in approximately 55% of all patients, in whom it most often presents as a sole aberration.5,10–13 A minimally deleted region of approximately 600kb has been described that contains a number of transcripts, including TRIM13, DLEU1 and 2, RCBTB1 and SETDB2, none of which has been shown to harbour any mutations or display altered expression in CLL.14–17 There is instead increasing evidence for the involvement of two microRNAs (mirs), mir-15a and mir-16-1, located in 13q14, in the pathogenesis of CLL.18 These mirs have been shown to negatively regulate the anti-apoptotic function of B-cell CLL/lymphoma 2 (BCL2).19,20 BCL2 is commonly found to be overexpressed in CLL,21,22 and defective apoptosis due to BCL2 overexpression has previously been suggested as an explanation for why tumour cells accumulate in CLL.23
Juliusson et al., on behalf of the International Working Party on Chromosomes in CLL (IWCCLL), were the first to show the prognostic significance of 13q14 deletions, where patients carrying this abnormality appeared to have a favourable prognosis.4,24 This finding has since been replicated in a number of studies, including a publication from our group.5,13 In addition, we detected a tendency among patients with a homozygous deletion of 13q, who account for approximately 20% of patients with del(13q), to have better clinical outcome compared with patients with heterozygous deletions only.13 This could partly be explained by a higher frequency of mutated immunoglobulin heavy-chain variable (IGHV) genes among patients with homozygous deletions (95%) compared with cases with only one deleted copy of 13q (73%).13 However, a recent paper found no significant difference in time to first treatment (TTT) or overall survival (OS) when studying patients with heterozygous and homozygous 13q deletions.25 Interestingly, the authors reported an association between the fraction of 13q- nuclei and clinical outcome, where patients carrying a high percentage of aberrant cells had a significantly shorter TTT, supporting similar findings in an earlier publication.26 This indicates that there is probably more to the 13q- story than previously believed, and warrants further characterisation.
Deletion of 11q22–q23
A deletion in the long arm of chromosome 11 is the second most common chromosomal abnormality in CLL, with a frequency that is currently estimated to be around 12–18% of all patients5,11,13 (see Figure 1). The minimal affected region is larger than that of the recurrent deletion in 13q and is mapped to a 2–3Mb region.27,28 There are several genes defined within this interval, with the ataxia telangiectasia mutated (ATM) gene being the most promising candidate. ATM is a key regulator of multiple signalling cascades that respond to DNA damage, and has been found to phosphorylate p53, thereby triggering the various p53-dependent signalling pathways.29 In addition, both deletion and missense mutations of ATM have been found in T-cell pro-lymphocytic leukaemia and mantle cell lymphoma.30–33 Mutations in ATM have been reported in CLL, where they are predominantly found in 11q-deleted patients.34–38
The prognostic relevance of 11q deletions in CLL was initially reported in the mid-1990s, when patients carrying this deletion were shown to have progressive disease and reduced survival.39–41 These findings have since been confirmed in numerous studies, including a recent publication from our laboratory.5,13 In multivariate analysis, the presence of 11q deletion is a significant predictor of clinical outcome.5 In addition, patients carrying an 11q deletion often display a distinctive clinical feature characterised by abdominal and mediastinal lymphadenopathy.41 Interestingly, there is recent evidence that chemoimmunotherapy including rituximab may overcome the adverse prognostic significance of 11q deletion in previously untreated patients with CLL.42
Trisomy 12
This recurrent chromosomal abnormality was the first and the most commonly detected aberration in the early days of CLL cytogenetics.1,43 Trisomy 12 is today detected in 11–16% of all patients with CLL,5,11,13 although an early study using FISH on interphase cells reported a frequency as high as 35%.44 A number of banding studies have identified a partial trisomy 12q in a few cases,45,46 which was subsequently narrowed down to 12q13–q15, and which contains the oncogenic murine double minute 2 (MDM2) gene.47,48 MDM2 functions as an important feedback regulator of TP53 and has been found to be dysregulated in CLL.49–51 Another candidate gene that has recently been in focus is the CLL upregulated gene 1 (CLLU1), which is located in 12q22.52 Interestingly, CLLU1 has been shown to display CLL-specific expression not detected in normal tissue or in other haematological malignancies. However, no increase in expression was found in patients carrying trisomy 12.52 Similarly, in a protein expression study of other chromosome 12 candidate genes, including CCND2, CDK2, CDK4 and STAT6, no difference in expression was demonstrated in cases with or without trisomy 12.53
Although the exact role of trisomy 12 in the pathogenesis of CLL is unclear, it has nevertheless been shown to have clinical significance. This was initially published at the beginning of 1990s by the IWCCLL, where patients carrying this abnormality were found to have the shortest survival among all patients with single aberrations.4,24 Today, trisomy 12 is associated with an intermediate survival and short TTT.5,13
Deletion of 17p
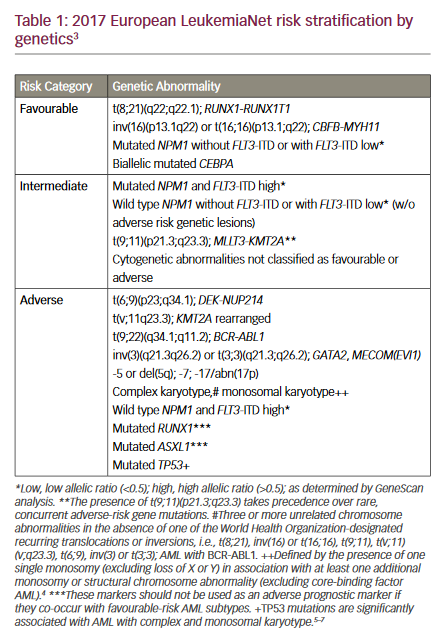
Loss of 17p, which harbours the TP53 gene, is detected in up to 10% of CLL patients, although a higher frequency is reported in fludarabine-refractory cases.5,11,13,54 In addition, the majority of patients displaying a 17p deletion also carry mutations on the remaining allele, whereas only up to 5% of non-17p-deleted cases are TP53– mutated.55–57 In CLL, loss of 17p is associated with the worst survival and shortest TTT, as well as treatment failure with purine nucleoside analogues and alkylating agents.5,13,58–62 Recent reports have also shown an independent prognostic value of TP53 mutations without the accompanying deletion.55,63 In addition, TP53 abnormalities are further associated with disease transformation to diffuse large B-cell lymphoma (Richter’s syndrome).64
The current agents used in chemotherapy aim to interrupt the cell cycle or induce apoptosis through the p53 pathway, which in this group of patients is rendered non-functional.65 Treatment with high-dose methylprednisolone, a glucocorticoid that acts independently of the p53 pathway, has been shown to be effective in patients with p53 defects.66 In addition, alternative treatment with the anti-CD52 monoclonal antibody alemtuzumab, which acts via complement-dependent cytotoxicity and antibody-dependent cell-mediated cytotoxicity, has shown promising results.67–69 Finally, refractory patients who are deemed to be physically fit, particularly younger patients, may be considered for allogeneic stem cell transplantation.70
In multivariate analysis of prognostic markers in CLL, deletion of 17p has repeatedly been shown to be one of the strongest independent markers of clinical outcome.6,7 Loss of 17p is most commonly associated with other poor prognostic markers, such as unmutated IGHV genes and advanced Binet stage.6 However, a recent study by Best et al. reported a subset of CLL patients with TP53 abnormalities and mutated IGHV genes having stable disease for several years without any need for therapy.71 We have found similar results in our laboratory (unpublished data). Thus, 17p abnormalities do not necessarily result in poor prognosis, and there seems to be a subgroup of patients who do not require treatment. This finding may argue against early introduction of chemotherapy in 17p-deleted patients without any clinical symptoms.
Deletions of 6q
Structural alterations in the long arm of chromosome 6 are commonly found in lymphoid malignancies.72,73 In CLL, a minimally deleted region has been defined in 6q21, and is reported in 6–7% of cases.74,75 In addition, a second recurrent deletion has been shown in 6q27.75,76 So far, no genes involved in the pathogenesis of CLL have been indicated for this region; however, absent in melanoma 1 (AIM1) has been suggested as a candidate gene.12,74,77 Although several studies showed deletions in 6q to be associated with higher white blood cell counts and more extensive lymphadenopathy, which may serve as a marker of progression, the prognostic value of this structural aberration remains unclear due to conflicting reports.75,78,79 We did not detect any deletions in 6q in a cohort of newly diagnosed CLL patients, which argues for this deletion as a marker of progression.13
Translocations Involving the Immunoglobulin Heavy-chain Locus
Recent advances in cultivation techniques, including stimulation of CLL cells with CpG oligonucleotides, interleukin 2 (IL-2) and CD40L, have resulted in an improved ability to detect chromosomal aberrations using conventional cytogenetics.80–82 As a result, chromosomal translocations that previously were thought to be rare events in CLL are now detected in more than 30% of patients.11,83,84 In addition, the presence of translocations, both balanced and unbalanced, was associated with poor clinical outcome.83,85 However, translocations are detected primarily as non-recurrent events. The most commonly affected region (detected in 27% of all translocations and 5% of all cases) involves the immunoglobulin heavy-chain (IGH) locus on 14q32.11 Chromosomal translocations involving the IG loci are found in many B-cell malignancies. These chromosomal rearrangements led to the dysregulation of oncogenes by juxtaposition to the IG loci. In CLL, patients carrying translocations involving the IGH locus identify a disease subset with poor prognosis.86
t(14;18)(q32;q21)–BCL2
Although characteristic of follicular lymphoma (FL), where this translocation involving BCL2 is observed in approximately 90% of cases, this rearrangement has also been shown in CLL.83,87,88 BCL2 is overexpressed in several lymphoid malignancies, including CLL.21,22,89 Interestingly, in FL the breakpoint in chromosome 18 occurs primarily at the 3’ end of BCL2, while in CLL the break tends to occur at the 5’ end of the same gene, juxtaposing BCL2 to the IG light chains.77
t(14;19)(q32;q13)–BCL3
The translocation involving the IGH and the BCL3 gene loci are present in various B-cell malignancies. They are most commonly found in CLL, where they are also associated with poor prognostic markers such as additional chromosomal changes, including trisomy 12 and unmutated IGHV genes.90 In addition, this translocation has been reported primarily in patients carrying IGHV4-39/IGHD6-13/IGHJ5 rearrangements.90
Novel Recurrent Aberrations
The application of high-density single-nucleotide polymorphism (SNP) arrays for evaluation of genomic aberrations in CLL has in recent years resulted in the detection of several new recurrent abnormalities. These abnormalities include gain of 2p and 8q and deletion of 4p, 8p and 22q, although most of them are detected at varying frequencies and their clinical relevance remains unclear.10,13,91,92 Gain of 2p, harbouring the MYCN oncogene, was recently found in up to 28% of untreated Binet stage B/C patients.93 Importantly, this recurrent aberration is associated with increased MYCN messenger RNA (mRNA) expression, suggesting MYCN to have a pathogenic role in CLL.93 Interestingly, we recently reported a small subgroup of CLL patients (n=5) with a detected gain in 2p to have a concurrent deletion of 11q and poor clinical outcome, requiring therapy.13 Deletion of 22q was detected in 15% of cases analysed in a study by Gunn et al.92 However, the authors found no association between del(22q) and clinical outcome. A number of subsequent studies using an alternative array platform have failed to detect this abnormality, perhaps due to the difference in array coverage.10,13 Finally, recurrent copy-number-neutral loss of heterozygosity (LOH), covering large parts of chromosome 13q, has been reported in patients with homozygous deletion of 13q14, although the clinical implication of this finding is still unknown.13,94
Genomic Complexity
The prognostic importance of genomic complexity in CLL was initially recognised in the mid-1980s, when increasing numbers of chromosomal aberrations within the cell clone were significantly associated with a poorer prognosis.43,95 Using CpG oligonucleotide- and CD40L-stimulated CLL cells and conventional cytogenetics, this association was further validated by Mayr et al. in 2006.83 In addition, a recent publication utilising high-density SNP arrays showed an association between genomic complexity, including non-recurrent changes in DNA copy number, and time to first and second treatment in CLL, where patients with two or more genomic aberrations had a significantly poorer clinical outcome.96 Our own investigations using high-resolution SNP arrays have confirmed these findings. Furthermore, we detected that patients carrying deletions of 17p have a considerably higher frequency of larger genomic aberrations than do patients with other recurrent aberrations, according to the hierarchal model defined by Döhner et al.5,13 (see Figure 2). In fact, when investigating patients with four or more aberrations >5Mbp, 50% carried 17p deletions while the remaining cases all displayed deletions of 11q. In addition, more than 80% of cases with two to three copy-number alterations (CNAs) >5Mbp carried deletions of 11q, deletions of 17p or trisomy 12, while patients with no recurrent alterations or patients carrying 13q deletions mostly had zero to one CNAs >5Mbp, indicating an association between genomic complexity and more aggressive disease. Furthermore, our data indicated that non-recurrent genomic aberrations <1Mbp in size did not carry prognostic information in CLL.13 Thus, while genomic complexity is clinically relevant, FISH analysis of recurrent genomic aberrations would in most cases identify the poor-prognosis cases.
Concluding Remarks
Despite massive developments in the field of molecular genetics and molecular cytogenetics, the cause of CLL still remains unknown and, as yet, no single genetic event has been indicated in all cases. Nevertheless, great improvements have been made in our understanding of CLL cancer genetics. The introduction of genomic microarrays has provided additional data on genome-wide DNA CNAs; while informative, as yet this has not resulted in breakthrough discoveries. The next necessary step will be the implementation of whole-genome sequencing in CLL research. Using next-generation sequencing techniques, it is possible to study the cancer genome at a new level of resolution, identifying all somatic mutations. These will include not only structural changes, but also mutations occurring in protein-coding genes, non-coding RNA genes or functional elements. In addition, other possible causes of oncogenesis, such as the presence of DNA viruses, can be discovered. This type of information will generate new insights into the genetic profile that underlies CLL, and may also provide additional clues to the pathogenesis of CLL. ■